Fig. 7.
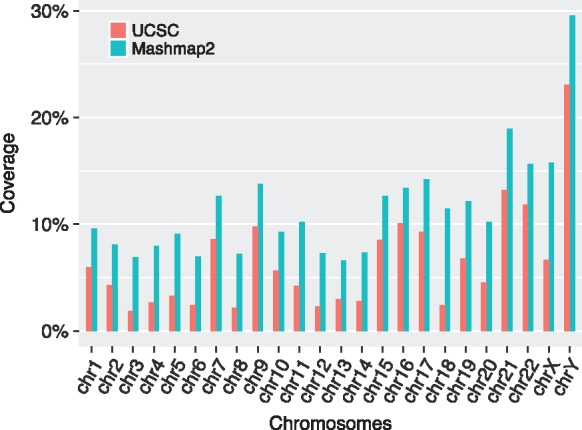
Comparison of genomic coverage between the UCSC Segmental Duplication database and Mashmap2 output alignments. Both methods reported equal coverage 83% on mitochondrial chromosome (not shown above to keep the plot legible). Coverage of duplications computed using our method is significantly higher, owing to its exhaustive search of all repeats with 1 Kbp length and 90% identity without repeat masking
