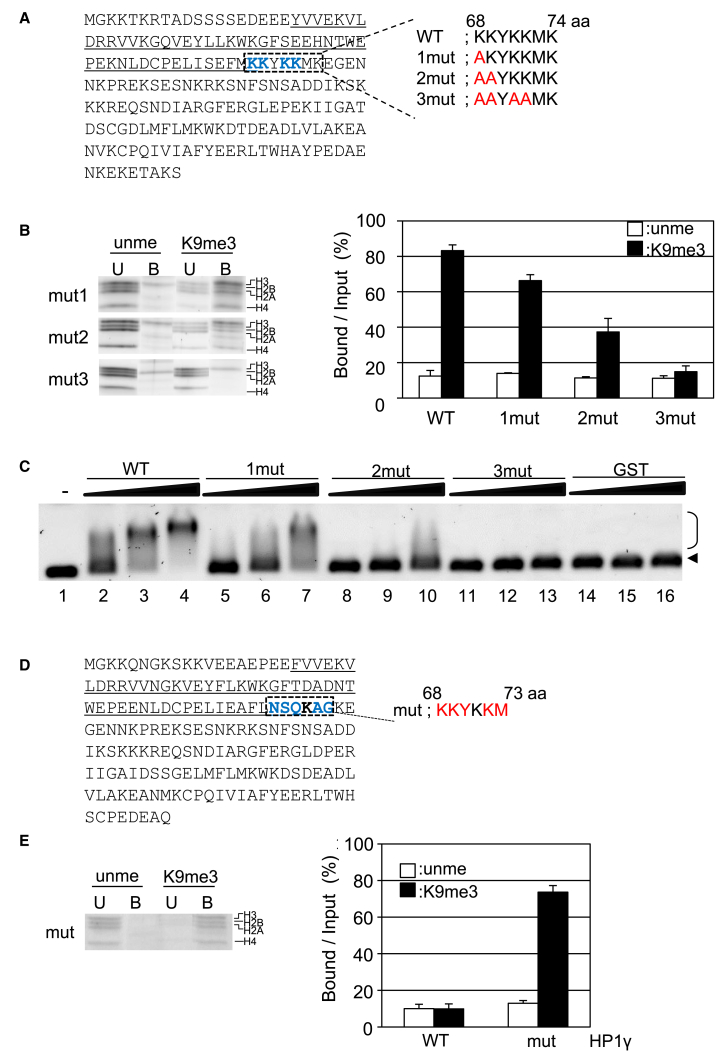
Figure 6.
Biochemical assays of HP1α mutations at basic regions in the hinge region. (A) The amino acid sequence of HP1α is given. The region corresponding to CD is underlined, and the mutation sites (K68, K69, K71, and K72) are shown as bold blue letters. The amino acid sequences of three mutants, 1mut (K68A), 2mut (K68A and K69A), and 3mut (K68A, K69A, K71A, and K72A) are also shown. (B) The binding activities of mutated HP1α to dinucleosomes are shown. The binding activity was examined as described in Fig. 2A. The activity of WT HP1α is taken from Fig. 2B. (C) The DNA binding activities of WT and mutated HP1α are shown. Naked DNA was incubated without protein (lane 1), with GST-WT HP1α (WT) (lanes 2–4), 1mut (lanes 5–7), 2mut (lanes 8–10), 3mut (lanes 11–13), or GST (lanes 14–16). To the DNA, 40 (lanes 2, 5, 8, 11, and 14), 80 (lanes 3, 6, 9, 12, and 15), or 160 (lanes 4, 7, 10, 13, and 16) molar excess of protein was added, and a gel retardation assay was performed. The arrowhead and bracket indicate the position of free DNA and shifted DNA, respectively. (D) The sequence of HP1γ is given. The CD is underlined, and the mutation sites are shown as bold blue letters. The mutated sequence is denoted in red. (E) Dinucleosomes reconstituted with unmethylated H3 (unme) or H3K9me3(K9me3) were mixed with the beads, and the binding assay was performed as described in Fig. 2B. Three unbound (U) and bound (B) fractions were separated and then subjected to SDS-PAGE, and each histone is indicated. The experiments were repeated three times, and the binding activity was summarized in the right panel. The binding activity of GST-HP1γ WT is taken from Fig. 2B. To see this figure in color, go online.