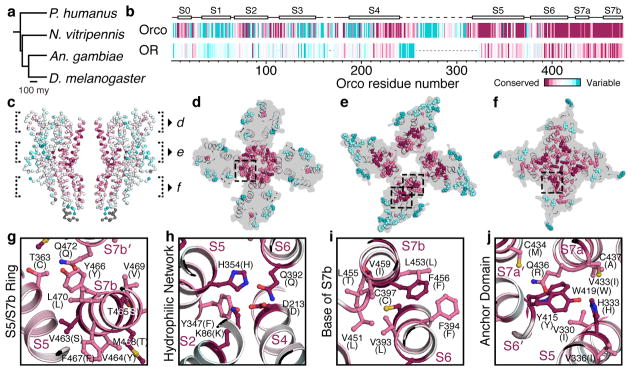
Fig. 5. OR conservation maps to key interaction domains in Orco.
a, Phylogenetic tree of insects whose OR sequences were used in the alignment. b, Residue conservation among 176 Orcos from 174 species (top) and 371 ORs from the species in a (bottom), calculated using ConSurf34, are plotted using a colour scale (far right) and aligned to A. bakeri Orco. Orco and OR conservation scores are independently normalized. Orco secondary structure is indicated above the plot. c, OR conservation scores from b mapped onto the structure of Orco. d–f, 15 Å cross-sections through c. The most conserved and variable amino acids are shown as spheres. g–j, Selected regions from d–f shown in greater detail. Residues are labeled according to A. bakeri Orco, with the most common OR amino acid at that position indicated in parentheses.