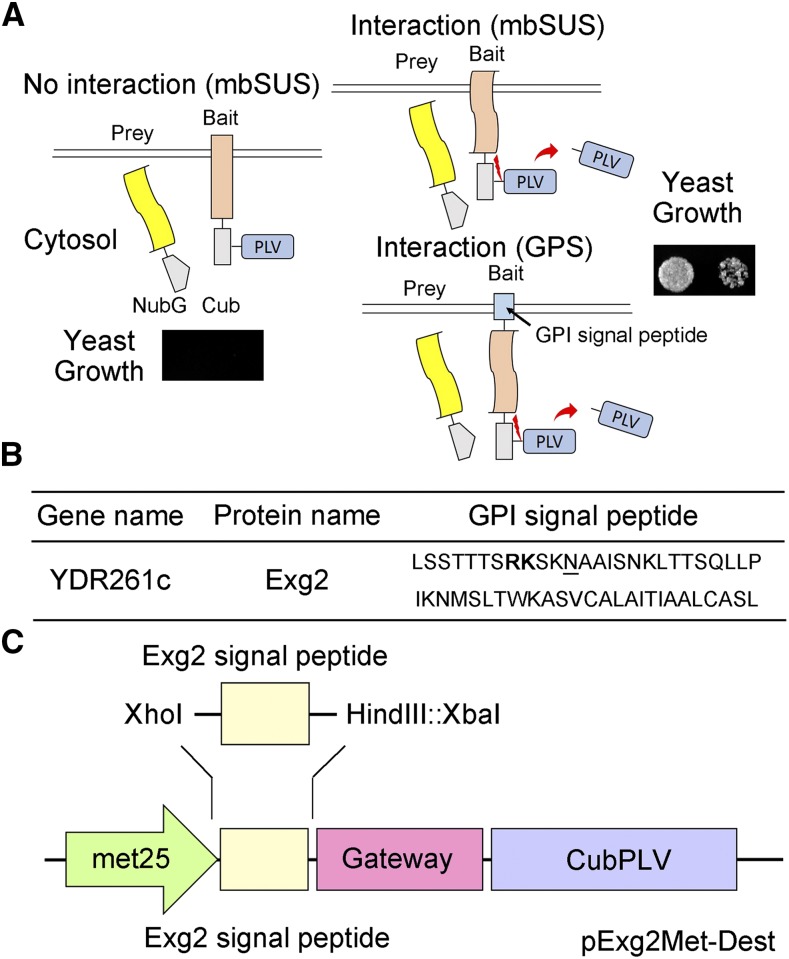
Figure 1.
The Mating Based Split-Ubiquitin System (mbSUS). A, Schematic of the split-ubiquitin system. As in conventional SUS methods, ubiquitin is split between the N-terminal half (Nub) and the C-terminal half (Cub), and the latter is fused with the transcriptional activator complex Protein A-LexA-VP16 (PLV). Mutating the isoleucine at position 13 of the wild-type Nub (NubI or NubWt) to Gly yields the NubG, which blocks spontaneous reassembly of ubiquitin. The NubG-Prey and Bait-CubPLV fusion constructs are transformed into mating strains of yeast that require differing amino-acid supplementation for growth. After mating, interaction of the two proteins expressed in the diploid yeast is tested by growth on selective media as in the standard SUS assay (Grefen et al., 2010; Zhang et al., 2015, 2017). Interaction between bait and prey reassembles a functional ubiquitin and leads to cleavage and release of the PLV complex by ubiquitin-specific proteases. Critical to this technology, the bait must be a membrane-bound protein, although the prey may be either membrane-bound or soluble. In the GPS system, the bait protein is fused at the N terminus with an Exg2 GPI signal peptide-anchor sequence, which anchors it in the membrane. B, The C-terminal GPI signal sequence of the Exg2 protein of yeast. The dibasic Arg-Lys (RK) residue motif is shown in bold, and the ω-site is underlined. C, Schematic of the pExg2Met-Dest vector used to express the bait fusion protein, including the N-terminal fusion with the GPI signal sequence (see also Supplemental Fig. S1).