Figure 1.
Rapid Nuclear Depletion of Mex67p Results in Global Inhibition of pA+ RNA Net Production
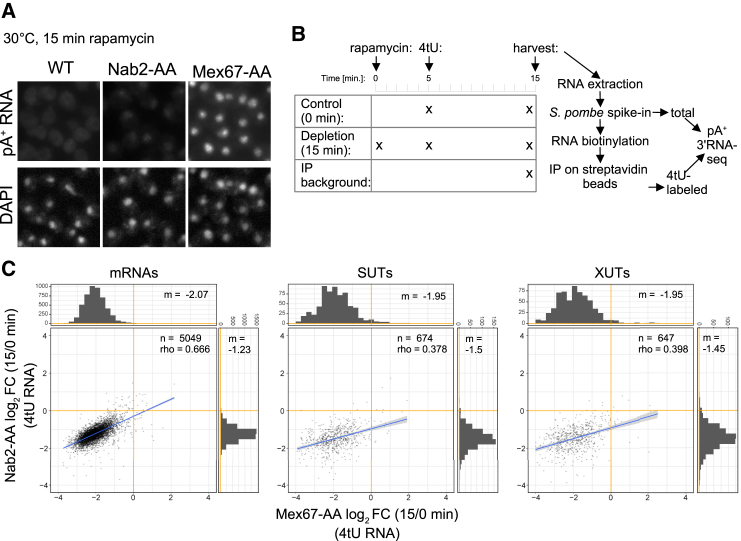
(A) Fluorescent in situ hybridization (FISH) analysis of pA+ RNA using a Cy3-labeled dT18 DNA/LNA probe on fixed WT, Nab2-AA, or Mex67-AA cells subjected to rapamycin treatment for 15 min at 30°C (top). FISH Cy3 images were adjusted to a common display range (defined as the highest and lowest pixel values for the three images) to allow comparison between strain phenotypes. DAPI staining was used to visualize chromatin-rich regions of cell nuclei (bottom).
(B) Schematic workflow of the pA+ RNA 3′ end sequencing experiment. Mex67-AA and Nab2-AA cultures were treated with 4tU for 10 min (“Control (0min)”) or pre-treated with rapamycin for 5 min before 4tU addition (“Depletion (15 min)”). Cells that were not treated with 4tU or rapamycin were used as reference to estimate the background of the 4tU purification (“IP background”). All obtained RNA samples were supplemented with S. pombe spike-in transcripts, biotinylated, and purified on streptavidin beads. Both total and 4tU-purified fractions were subjected to pA+ RNA 3′ end sequencing.
(C) Log2 fold changes (FC) of pA+ RNA net production in 4tU-labeled Mex67-AA (x axis) and Nab2-AA (y axis) cells treated with rapamycin for 15 min and compared with their respective controls (no rapamycin). Three major pA+-containing transcript classes were analyzed: mRNAs, SUTs, and XUTs. Only transcripts purified significantly above background in the control samples are depicted. The number of genes (n), Spearman correlation coefficients (rho), and median (m) values for both datasets are specified on each graph. See also Table S1.
See also Figure S1.