Figure 2.
Decreased pA+ RNA Net Production in Mex67p- and Nab2p-Depleted Cells Is Due to Increased Transcript Decay
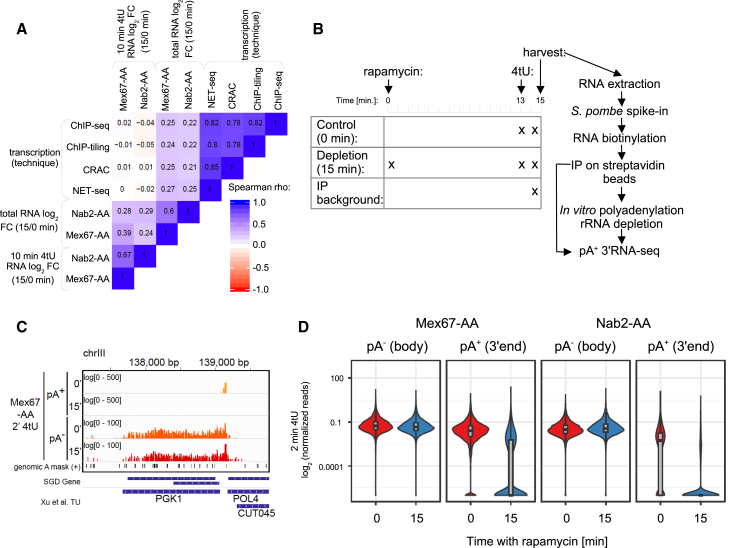
(A) Spearman correlation coefficient matrix comparing mRNA log2 FC of 15/0 min rapamycin ratios in Mex67- and Nab2-AA cells as in Figures 1C and S1C against various estimates of transcription rates. Transcription estimates were derived from RNAPII NET-seq (Churchman and Weissman 2011), RNAPII-CRAC (Milligan et al., 2016), RNAPII-ChIP-seq (Warfield et al., 2017), and RNAPII-ChIP-tiling (Mayer et al., 2010) data.
(B) Schematic workflow of the pA+/pA− RNA 3′ end sequencing experiment. Cells were treated with 4tU for 2 min. In case of the Mex67-AA- and Nab2-AA-depleted cells, cultures were pre-treated with rapamycin for 13 min before 4tU labeling. Addition of S. pombe spike-in RNAs, biotinylation, and purification of 4tU-labeled RNA were done as described in Figure 1B. Samples were then split in two aliquots, in which one was subjected to pA+ RNA 3′ end sequencing as in Figure 1B, and the other was in vitro polyadenylated and rRNA-depleted prior to pA+ RNA 3′ end sequencing.
(C) Genome Browser view of spike-in normalized pA− and pA+ RNA 3′ end reads derived from 2 min 4tU-labeling experiments of untreated (0′) and 15 min nuclear-depleted (15′) Mex67-AA cells, spanning across the gene-coding strand of PGK1 on chromosome III. Annotations below the tracks show genomic A-rich sites masked in the analysis and boundaries of translated and transcribed regions as previously defined in www.yeastgenome.org and Xu et al. (2009), respectively.
(D) Levels of nascent (“pA− (body)”: mean of pA− signal in the region from gene TSS to 200 bp upstream of TES) and mature (“pA+ (3′ end)”: mean of pA+ signal in the region from 200 bp up- and downstream of TES) mRNAs in 2 min 4tU-labeled samples in Mex67-AA (left) and Nab2-AA (right) cells after 0 (red violins) or 15 (blue violins) min of rapamycin treatment. All signals are shown relative to S. pombe spike-ins, with background subtracted, log2 and length scaled. Levels of transcripts that were not purified above background were set to a pseudocount of 10−6. Violin plots shown are overlaid with box plots depicting the median values with boxes demarcating first and third quartiles of the data. See also Table S2.
See also Figure S2.