Figure 2.
pA− RNA 3′ End Counts Correlate with Other Transcription Measures
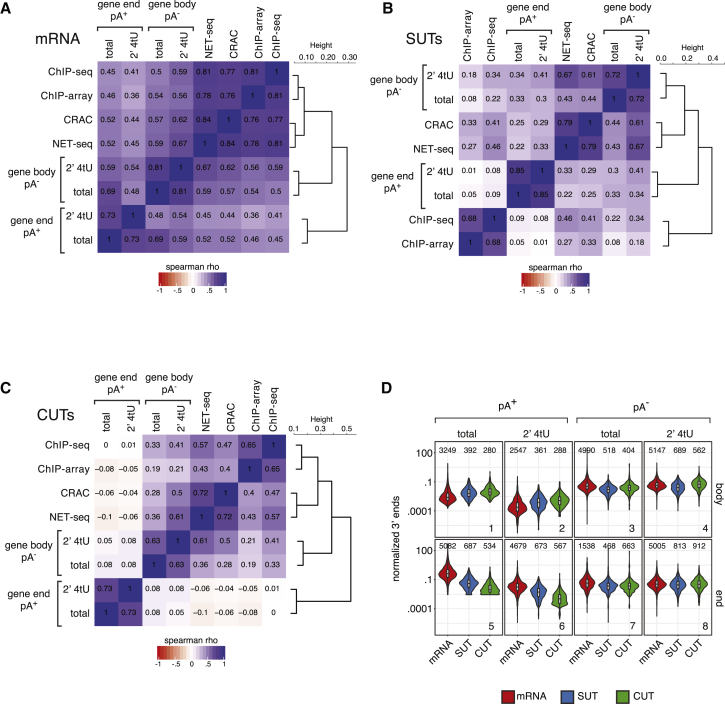
(A) Spearman rank correlation (rho) matrix and hierarchical clustering of pA− signal densities in gene body (TSS to 200 bp upstream of TES) and pA+ signals in gene end (TES ± 200 bp) regions of mRNAs (n = 5,171) and those reported from RNAPII ChIP-seq (Warfield et al., 2017), RNAPII ChIP-tiling array (ChIP-array; Mayer et al., 2010), NET-seq (Churchman and Weissman, 2011), and RNAPII CRAC (Milligan et al., 2016) datasets.
(B) As in (A), but for SUT TUs (n = 847).
(C) As in (A), but for CUT TUs (n = 925).
(D) Violin plots depicting the distribution of signal densities in gene body and end regions for pA+ (left) and pA− (right) signals. Values for mRNA, SUT, and CUT TUs are shown separately for total and 2′ 4tU samples. Number of TUs considered are depicted above each violin.
See also Figure S2.