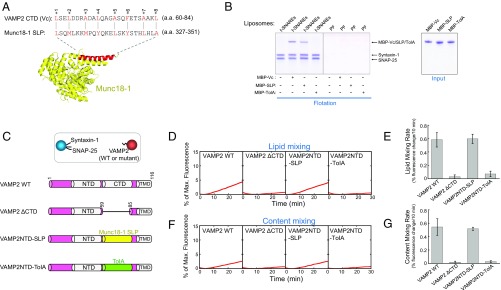
Fig. 4.
SM proteins possess a SNARE-like peptide that structurally and functionally resembles Vc peptide. (A, Top) Alignment of VAMP2 CTD (Vc peptide) and the SNARE-like peptide (SLP) from Munc18-1 (identity: 20.0%; similarity: 56.0%). The alignment was performed using the LALIGN program (https://embnet.vital-it.ch/software/LALIGN_form.html). Alignment method, global; scoring matrix, PAM250; other parameters, default. Layer residues of Vc peptide are numbered and highlighted. (A, Bottom) Crystal structure of Munc18-1 with SLP shown in red (PDB ID code 3PUJ) (27). (B) Coomassie blue-stained SDS/PAGE gels showing the binding of Vc peptide, Munc18-1–derived SLP, and the bacterial TolA helix to the indicated liposomes. The t-SNARE complex was composed of syntaxin-1 and SNAP-25. PF, protein free. Each binding reaction contained 5 μM SNAREs and 5 μM MBP-tagged peptides. (C, Top) Illustration of the liposome fusion pairs. (C, Bottom) Diagrams of WT and mutant VAMP2 proteins used in the liposome fusion assays. The sequences of the chimeras are included in Materials and Methods. (D) Lipid mixing of the reconstituted liposome fusion reactions containing WT synaptic exocytic t-SNAREs and WT or mutant VAMP2. Each fusion reaction contained 5 μM t-SNAREs, 1.5 μM v-SNARE, and 100 mg/mL Ficoll 70. (E) Initial rates of the lipid mixing reactions shown in D. (F) Content mixing of the liposome fusion reactions described in D. (G) Initial rates of the lipid mixing reactions shown in F. Data in E and G are presented as percentage of fluorescence change per 10 min. Error bars indicate SD.