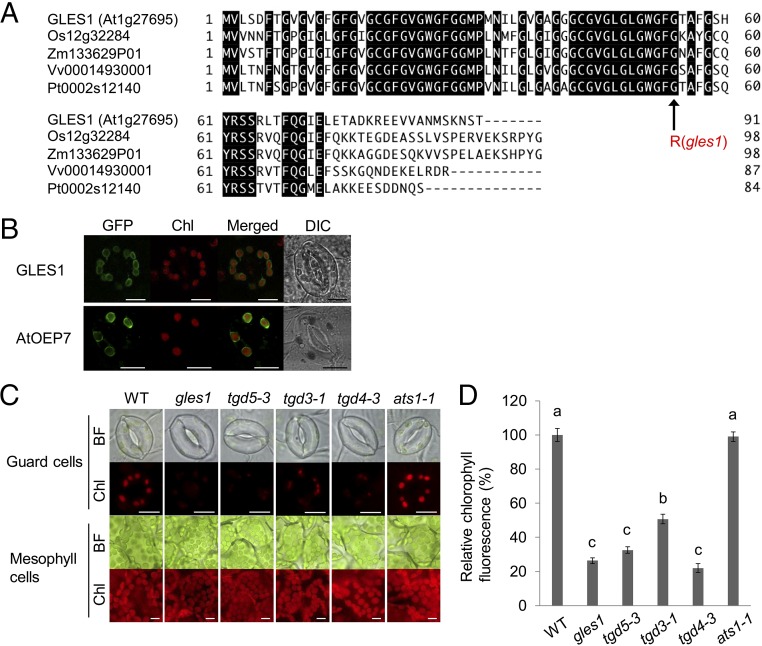
Fig. 2.
TGD proteins involved in lipid transport from ER to chloroplasts are essential for guard cell chloroplast development. (A) Alignment of amino acid sequence of the GLES1(At1g27695) protein and its orthologs assessed using Clustal W. GLES1-like proteins are found in various plants, including Oryza sativa (Os12g32284), Zea mays (Zm133629P01), Vitis vinifera (Vv00014930001) and Populus trichocarpa (Pt0002s12140). Amino acid identity is indicated by black boxes. The gles1 mutation site Glycine 53 is conserved among the orthologs, as indicated by the arrowhead. (B) Chloroplast envelope localization of GLES1-GFP. Expression of GLES1–GFP fusion protein resulted in functional complementation of the gles1 mutant phenotypes (Fig. 1 A and B). GFP fluorescence, autofluorescence of chlorophyll, the overlay of all fluorescence signals, and the differential interference contrast images (DIC) are shown for a representative example. AtOEP7-GFP was used as the chloroplast envelope marker. Chl, Chlorophyll fluorescence. (Scale bars, 10 μm.) (C and D) GLES1 is identical to TGD5, a subunit of TGD protein involved in the ER-to-plastid transport. The T-DNA insertion allele tgd5-3 and loss of function mutants of other TGD complex subunits exhibited achlorophyllous phenotypes in guard cells. (Scale bars, 10 μm.) Total chlorophyll autofluorescence of individual guard cells was analyzed (D). The average chlorophyll autofluorescence measured for WT plants was designated as 100%, and relative fluorescence has been plotted (%). Values shown are means ± SE (n > 100) of five independent experiments. The statistical significance was determined by a one-way ANOVA with Tukey-Kramer multiple comparison tests. Same letters (a–c) indicate no significant difference (P > 0.05).