Significance
Chimpanzees, bonobos, and gorillas harbor close relatives of human Plasmodium vivax, but current knowledge of these parasites is limited to a small number of gene fragments derived almost exclusively from mitochondrial DNA. We compared nearly full-length genomes of ape parasites with a global sample of human P. vivax and tested the function of human and ape P. vivax proteins believed to be important for erythrocyte binding. The results showed that ape parasites are 10-fold more diverse than human P. vivax and exhibit no evidence of species specificity, whereas human P. vivax represents a bottlenecked lineage that emerged from within this parasite group. Thus, African apes represent a large P. vivax reservoir whose impact on human malaria eradication requires careful monitoring.
Keywords: Plasmodium vivax, genomics, malaria, great apes, zoonotic transmission
Abstract
Wild-living African apes are endemically infected with parasites that are closely related to human Plasmodium vivax, a leading cause of malaria outside Africa. This finding suggests that the origin of P. vivax was in Africa, even though the parasite is now rare in humans there. To elucidate the emergence of human P. vivax and its relationship to the ape parasites, we analyzed genome sequence data of P. vivax strains infecting six chimpanzees and one gorilla from Cameroon, Gabon, and Côte d’Ivoire. We found that ape and human parasites share nearly identical core genomes, differing by only 2% of coding sequences. However, compared with the ape parasites, human strains of P. vivax exhibit about 10-fold less diversity and have a relative excess of nonsynonymous nucleotide polymorphisms, with site-frequency spectra suggesting they are subject to greatly relaxed purifying selection. These data suggest that human P. vivax has undergone an extreme bottleneck, followed by rapid population expansion. Investigating potential host-specificity determinants, we found that ape P. vivax parasites encode intact orthologs of three reticulocyte-binding protein genes (rbp2d, rbp2e, and rbp3), which are pseudogenes in all human P. vivax strains. However, binding studies of recombinant RBP2e and RBP3 proteins to human, chimpanzee, and gorilla erythrocytes revealed no evidence of host-specific barriers to red blood cell invasion. These data suggest that, from an ancient stock of P. vivax parasites capable of infecting both humans and apes, a severely bottlenecked lineage emerged out of Africa and underwent rapid population growth as it spread globally.
The protozoal parasite Plasmodium vivax causes over 8 million cases of human malaria per year, with the great majority occurring in Southeast Asia and South America (1). P. vivax is rare in humans in Africa due to the high prevalence of the Duffy-negative mutation (2), which abrogates expression of the Duffy antigen receptor for chemokines (DARC) on erythrocytes. Since DARC serves as a receptor for P. vivax, its absence protects Duffy-negative humans from P. vivax infection (3), although this protection is not absolute (4). Until recently, P. vivax was thought to have emerged in Asia following the cross-species transmission of a macaque parasite (5, 6). However, the finding of closely related parasites in wild-living chimpanzees and gorillas suggested an African origin of P. vivax (7). Indeed, parasite sequences closely resembling P. vivax have been detected in western (Pan troglodytes verus), central (Pan troglodytes troglodytes), and eastern (Pan troglodytes schweinfurthii) chimpanzees, eastern (Gorilla beringei graueri) and western lowland (Gorilla gorilla gorilla) gorillas, and most recently in bonobos (Pan paniscus) (7–11). Phylogenetic analyses of available sequences revealed that ape and human parasites were nearly identical, with human P. vivax sequences forming a monophyletic lineage that usually fell within the radiation of the ape parasites (7). These findings suggested that P. vivax infected apes, including humans, in Africa, until the spread of the Duffy-negative mutation largely eliminated the parasite in humans there. However, definitive conclusions could not be drawn, since all analyses of ape P. vivax genomes to date have rested on a small number of gene fragments amplified almost exclusively from parasite mitochondrial DNA present in ape fecal samples.
Understanding the origin of human P. vivax and its relationship to the ape parasites is important for several reasons. First, only six Plasmodium species, out of several hundred so far described as infecting vertebrate hosts (12), have successfully colonized humans (13). Thus, the circumstances that surround the emergence of each of these human pathogens are of interest, especially since most, if not all, have nonhuman primate parasites as their closest relatives (7, 9, 14). Second, it is currently unclear whether the ape parasites represent a separate species distinct from P. vivax. Although sequences from human P. vivax parasites form a monophyletic clade in phylogenetic trees, this may reflect their geographic separation and not the existence of host-specific infection barriers. Indeed, ape P. vivax has been shown to cause malaria in a Duffy-positive European traveler (11), and human P. vivax has been transmitted to wild-living monkeys in South America, generating what has been called “Plasmodium simium” (15). If cross-species infection and recombination of ape and human P. vivax are possible, as appears to be the case for P. simium and P. vivax in South America (15, 16), this could have implications for malaria-eradication efforts. Finally, ape and human P. vivax strains may have acquired adaptations that limit parasite transmission between different host species. Such findings could explain why the macaque parasites Plasmodium knowlesi and Plasmodium cynomolgi can infect and cause malaria in humans but do not appear to be commonly transmitted between different human hosts (17).
To elucidate the events that led to the emergence of human P. vivax, we sought to obtain genome sequences of parasites infecting chimpanzees and gorillas. A similar approach has recently uncovered processes that may have allowed the gorilla precursor of Plasmodium falciparum to cross the species barrier to infect humans (18, 19). However, obtaining blood samples from Plasmodium-infected apes is challenging due to the endangered species status of these hosts. Moreover, ape P. vivax, like its human-infecting counterpart, exhibits only low levels of parasitemia (7) and has not been cultured. Although removal of host leukocytes from whole-blood samples (20) and parasite nucleic acid capture (21) have improved the recovery of human P. vivax genomes, these approaches are not readily applicable to ape parasites. We thus adapted a previously developed selective whole-genome amplification (SWGA) method (18) to generate P. vivax genome sequences from unprocessed chimpanzee and gorilla blood samples obtained in different parts of Africa. Analysis of these genomes revealed that the ape parasites are about 10 times more diverse than global representatives of human P. vivax (21), indicating that the human parasite has undergone a severe genetic bottleneck. Ape P. vivax genomes were found to have intact orthologs of three reticulocyte-binding protein (RBP) genes that are pseudogenized in all human P. vivax strains, but functional studies of two of the encoded proteins revealed no evidence of species-specific receptor interactions. The P. vivax ancestor therefore likely infected both humans and apes in Africa before being eliminated in humans there by the spread of the Duffy-negative mutation, while the current human-infecting parasites represent a lineage that had escaped out of Africa.
Results
Genome Assemblies of Chimpanzee P. vivax.
Leftover blood samples from routine health examinations of chimpanzees cared for at the Sanaga-Yong (SY) Chimpanzee Rescue Center in Cameroon were screened for Plasmodium infection using nested PCR with pan-Plasmodium and P. vivax-specific primers. Two samples, SY56 and SY43, were positive for ape P. vivax, with limiting-dilution PCR detecting one strain in SY56 and up to five variants in SY43, with two strains predominating. Since these two samples lacked other Plasmodium species, they were suitable for SWGA without the risk of generating interspecies recombinants (18, 22). SWGA uses the highly processive phi29 DNA polymerase and specific primers to preferentially amplify pathogen sequences from complex mixtures of target and host DNA and has been used successfully in the past to generate Plasmodium sequences from blood smear negative, unprocessed blood samples (18). Since SWGA can result in stochastic amplification when target templates are rare (18, 23), each sample was amplified on more than one occasion using different primer sets, with and without digestion of sample DNA with methylation-dependent restriction enzymes to degrade host DNA (SI Appendix, Table S1). Individual SWGA reactions were pooled and sequenced on Illumina and PacBio platforms (SI Appendix).
Draft genomes of the chimpanzee P. vivax strains PvSY56 and PvSY43 were generated using iterative reference-guided assembly to the human PvP01 reference genome (24) followed by gap-filling steps. In addition, PvSY56 reads that did not map to the PvP01 core genome were de novo assembled to obtain subtelomeric contigs. The resulting assemblies yielded 21.9 Mbp and 21.2 Mbp of sequence for PvSY56 and PvSY43, respectively (Table 1). Because sample SY43 contained at least five P. vivax strains (7), the PvSY43 genome represents a consensus of these variants. Annotations were transferred from PvP01, with additional genes predicted in the de novo contigs. Since a large number of genes contained frameshifts in homopolymer tracts, we manually corrected annotations spanning these presumed sequencing errors to maintain an ORF. Overall, PvSY56 and PvSY43 shared a highly conserved core genome with human P. vivax. More than 98% of PvP01 core genes (as defined in ref. 20) were identified in each chimpanzee P. vivax assembly (Table 1), with 88% present as full-length genes. Although 10 human P. vivax core genes were absent from both PvSY56 and PvSY43, ape P. vivax reads at least partially covered these coding regions, implicating assembly difficulties rather than differences between ape and human P. vivax genomes as the reason for their absence. Assembly issues likely also account for the small number of genes in subtelomeric and internal hypervariable regions that could be annotated for PvSY56 and PvSY43 (Table 1).
Table 1.
Genome features of ape P. vivax
| Genome attributes | PvSY56 | PvSY43* | PvP01† |
| Host species | Chimpanzee | Chimpanzee | Human |
| Country | Cameroon | Cameroon | Indonesia |
| Chromosomal assembly‡, bp | 21,928,114 | 21,224,756 | 24,177,188 |
| Mean depth of coverage§ | 319 | 240 | N/A |
| Chromosomal contigs | 7,112 | 6,604 | 14 |
| G + C content, % | 44.1 | 44.6 | 43.3 |
| Core protein-coding genes¶ (% of PvP01) | 4,883 (98.8) | 4,908 (99.3) | 4,941 (100) |
| Full length# (% of PvP01) | 4,391 (88.9) | 4,350 (88.0) | N/A |
| Partial (% of PvP01) | 492 (10.0) | 558 (11.3) | N/A |
| Genes in hypervariable regions¶ | 415 | 276 | 1,702 |
N/A, not applicable.
The genome assembly of PvSY43 represents a consensus sequence of at least two major and three minor chimpanzee P. vivax variants (Fig. 2 C and D and SI Appendix, Fig. S5).
Chimpanzee P. vivax genomes were compared with the human P. vivax reference PvP01 (24).
Number of unambiguous bases.
Calculated by dividing the number of nucleotides in reads mapped to the assemblies by the expected genome size from PvP01.
Subtelomeric, core and internal hypervariable regions were defined as described (20).
Genes classified as full length comprised at least 90% of the length of the corresponding PvP01 ortholog.
Polymorphism in Ape and Human P. vivax.
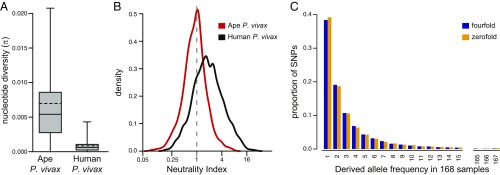
Comparison of coding sequences between the PvSY56 and PvSY43 assemblies revealed that they differ at 0.61% of sites, in contrast with a difference of only 0.11% between the two human P. vivax reference genomes, PvSalI and PvP01 (SI Appendix, Fig. S1A). Since PvSY56 and PvSY43 were derived from chimpanzees housed at the same sanctuary, we reasoned that they might not represent the full extent of ape P. vivax diversity (the two human reference strains were sampled on two different continents, in Latin America and Southeast Asia). This prompted us to obtain P. vivax genome sequences from additional infected apes. Using SWGA followed by Illumina sequencing, we amplified ape P. vivax from blood samples of two additional SY chimpanzees (SY81 and SY90), from a wild-living western chimpanzee (Sagu) from Côte d’Ivoire (10), and from a western lowland gorilla (Gor3157) sampled in Cameroon (SI Appendix, Table S2). We also mined the read database from a blood sample of a Plasmodium malariae-infected sanctuary chimpanzee from Gabon (14), which we had noted contained a substantial number of ape P. vivax reads. Reads from each sample were mapped to the PvSY56 assembly, and SNPs were identified. The extent of genome coverage varied considerably among the six chimpanzee samples; however, we were able to recover between 695 and 3,005 core genes (SI Appendix, Table S2), with 65% of genes analyzed being covered in four or more parasite genomes (SI Appendix, Fig. S2). The gorilla P. vivax strain, which was derived from a partially degraded bushmeat sample, yielded only 10 genes despite repeated amplification and thus was not included in the diversity analysis. For comparison, we included sequences from seven additional human P. vivax strains (21), each from a different country (India, Myanmar, Papua New Guinea, Thailand, Colombia, Mexico, and Peru), and identified SNPs using the same methods. Diversity values were then calculated across a common set of 4,263 core genes for which we obtained sequences from two or more strains for both ape and human P. vivax (Table 2). The results revealed a mean pairwise nucleotide sequence diversity (π) among the six chimpanzee P. vivax strains of 0.698%, about eight times higher than the value (0.085%) for the global sample of human strains (Fig. 1A, Table 2, and SI Appendix, Fig. S1B). Removal of the PvSY43 sequence did not decrease this difference (SI Appendix, Fig. S1C), indicating that the inclusion of this multiply infected sample did not inflate the diversity of the chimpanzee P. vivax parasites. Furthermore, analysis of transition/transversion ratios at fourfold degenerate sites yielded nearly identical results for chimpanzee (1.08) and human (1.07) P. vivax strains, excluding the possibility of an SWGA-related increase in diversity. Thus, the much higher level of diversity among chimpanzee P. vivax strains, compared with parasites currently circulating in humans, does not appear to be an artifact.
Table 2.
Nucleotide polymorphism in ape and human P. vivax
| Parasites | n* | πall† | π0‡ | π4§ | NS polymorphisms¶ | S polymorphisms¶ | NS/S |
| Ape P. vivax | 6 | 0.00698 | 0.00357 | 0.01604 | 32,364 | 47,494 | 0.68 |
| Human P. vivax | 9 | 0.00085 | 0.00060 | 0.00143 | 10,530 | 7,673 | 1.37 |
n = number of strains included in the analysis (see SI Appendix, Fig. S2 for gene coverage among the different strains).
Mean pairwise diversity at coding sites from 4,263 genes (6.5 million sites).
Mean pairwise diversity at zerofold degenerate sites from 4,263 genes (4.0 million sites).
Mean pairwise diversity at fourfold degenerate sites from 4,263 genes (0.7 million sites).
Numbers of nonsynonymous (NS) and synonymous (S) polymorphisms were calculated by counting the number of SNPs that changed (NS) or did not change (S) the protein sequence of the respective reference (PvSY56 for ape P. vivax; PvP01 for human P. vivax).
Fig. 1.
Nucleotide sequence diversity in ape and human P. vivax. (A) The π calculated across a common set of 4,260 core genes for six chimpanzee and nine human P. vivax strains (as in Table 2, but three genes with fewer than 35 aligned sites were excluded). Median and mean (weighted by gene length) π values are indicated by solid and dashed lines, respectively; box and whiskers indicate the interquartile range and 99th percentiles, respectively. Plots including outliers are shown in SI Appendix, Fig. S1B. (B) Density plots of NI values shown on a log2 scale for ape (red) and human (black) P. vivax genes. Values are shown for 1,585 genes with nonzero values of NI in both populations. (C) Site-frequency spectra of polymorphisms at fourfold degenerate (blue) and zerofold degenerate (orange) sites extracted from SNP data of human P. vivax samples from Southeast Asia (20).
The nature of the nucleotide polymorphisms also differed substantially between chimpanzee and human P. vivax strains. Among the chimpanzee parasites the majority of polymorphisms were synonymous, with the ratio of nonsynonymous to synonymous SNPs (NS/S) being 0.68. In contrast, the majority of polymorphisms among the human strains were nonsynonymous, with an NS/S ratio of 1.37 (Table 2). The NS/S ratio among polymorphisms can be compared with (i.e., divided by) the NS/S ratio among interspecies differences to yield the neutrality index (NI) (25). This NI assumes that synonymous changes, both within and between species, are selectively neutral and has an expected value of one when nonsynonymous changes are also neutral. We thus compiled a set of 3,913 genes that were comparable among ape and human P. vivax strains as well as between these parasites and the macaque parasite P. cynomolgi (their closest relative with an available genome sequence). The overall NI value for ape P. vivax strains was close to the neutral expectation (NI = 0.96) (SI Appendix, Table S3). In contrast, the overall value for human P. vivax strains was much larger, NI = 1.91, indicating a large excess of nonsynonymous polymorphisms among human strains relative to the expectation derived from patterns of divergence between species.
Comparison of the ratios of nonsynonymous and synonymous changes between within-species polymorphisms and between-species fixed differences forms the basis of the McDonald–Kreitman (MK) test for adaptive evolution of individual genes (26). The discrepancy in overall NI values for ape versus human P. vivax strains indicates that MK tests are likely to produce different results depending on which P. vivax strains are used. For the chimpanzee P. vivax strains, only two genes yielded significant results after correction for multiple testing (P < 0.05), both with NI <1 indicating a significant excess of fixed, potentially adaptive, nonsynonymous differences: one was found to be orthologous to PVP01_1201800, which is an immunogenic member of the tryptophan-rich antigen family of P. vivax (27, 28), and the other (orthologous to PVP01_1406200) encodes a conserved Plasmodium protein of unknown function. For the human P. vivax strains, five genes yielded significant MK test results, but all with NI >1, indicating an excess of nonsynonymous polymorphisms (Dataset S1). Such results are usually interpreted as evidence of balancing selection maintaining polymorphism, but the large overall NI value for the human strains suggests that a more pervasive factor, such as past demography, is influencing these genes. Fig. 1B shows the distributions of NI values for 1,585 individual genes with nonzero values in both ape and human P. vivax. These results indicate that the difference between ape and human P. vivax is not due to a subset of unusual genes but rather that the entire distribution is shifted from being centered around 1.0 in ape P. vivax to being centered around 2.0 in human P. vivax (Fig. 1B and Dataset S1).
To look for possible human-specific adaptive changes, we also performed MK tests for 4,263 core genes comparing polymorphisms within ape P. vivax with fixed differences between ape and human P. vivax. After correcting for multiple tests, we found no genes with a significant excess of nonsynonymous fixed differences. While this may seem surprising, the timespan since the human lineage of P. vivax became restricted to this host may have been too short for the accumulation of a sufficient number of adaptive changes to yield significant test results, and this finding does not exclude the possibility that smaller numbers of adaptive changes have indeed occurred.
To investigate the unusual pattern of nucleotide polymorphisms in human P. vivax in greater detail, we examined the frequencies of nonsynonymous and synonymous polymorphisms per site. We obtained different results depending on the methodology used, primarily because different methods use different approaches to estimate the numbers of sites available for synonymous changes. To avoid this problem, we included only fourfold and zerofold degenerate sites where, due to the structure of the genetic code, either all or none of the possible changes are synonymous. Among ape P. vivax strains, the nucleotide diversity at zerofold degenerate sites (0.357%) was 22% of that at fourfold degenerate sites (1.604%), whereas among human P. vivax strains, the value at zerofold degenerate sites (0.060%) was 42% of that at fourfold degenerate sites (0.143%) (Table 2 and SI Appendix, Fig. S1 D and E). Thus, while chimpanzee parasites were 11 times more diverse than human parasites at fourfold degenerate sites, they were only six times more diverse at zerofold degenerate sites, indicating that nonsynonymous polymorphisms in human P. vivax strains are almost twice as numerous as expected.
A large number of human P. vivax genome sequences have been characterized (20, 21), so it is possible to investigate the frequencies at which SNPs are segregating within the population. Synonymous polymorphisms are expected to be neutral, so their site frequency spectrum (SFS) should reflect past demography. By comparison, many nonsynonymous polymorphisms are expected to be slightly deleterious, and their SFS thus should be more skewed toward lower frequencies (29). To examine a large number of parasite sequences from a single geographic region, we focused on SNP data from Malaria Genomic Epidemiology Network (MalariaGen) samples from Southeast Asia (20). Ancestral and derived alleles at each site were identified by comparison with two outgroups: the chimpanzee P. vivax strain PvSY56 and a P. cynomolgi reference strain. The unfolded site frequency spectra obtained for SNPs at zerofold and fourfold degenerate sites are almost identical (Fig. 1C). Thus, the unusually large fraction of nonsynonymous polymorphisms among human P. vivax sequences appears to reflect mutations that are segregating as effectively neutral alleles.
Relationship of P. vivax Strains from Humans and Apes.
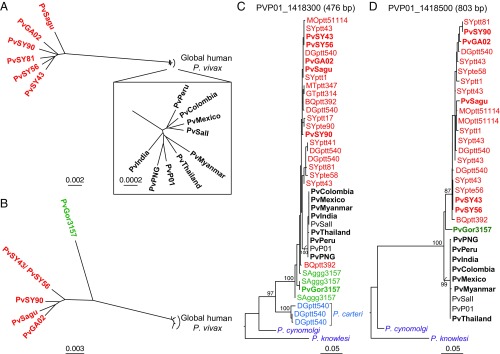
In previous analyses of a small number of partial gene sequences, we found that sequences from human P. vivax parasites always formed a monophyletic clade, which usually fell within the radiation of sequences from chimpanzee and gorilla samples (7). This was observed for mitochondrial and apicoplast sequences as well as for three nuclear genes, while for a fourth nuclear gene the ape and human P. vivax sequences formed sister clades (7). Here we found that, across their genome, chimpanzee parasites were much more divergent from the human parasites than they were from each other. For example, across 3,958 core genes, the chimpanzee parasite genomes PvSY56 and PvSY43 differed from one another at 0.6% of sites but differed from the human P. vivax reference genomes PvSalI and PvP01 at 2.2% of sites on average. This relationship is summarized in a neighbor-joining tree constructed from a matrix of pairwise genetic distances from an alignment of 241 nuclear genes available for all six chimpanzee parasites (Fig. 2A). Although this tree may not reflect the true evolutionary history of any one particular gene (due to recombination), the overall relationships were confirmed in a phylogenetic network (SI Appendix, Fig. S3A), which showed that the chimpanzee parasites sampled at the same location (SY) in Cameroon are on average a little more closely related to each other than they are to the strains identified in Gabon (GA02) and Côte d’Ivoire (Sagu). Inclusion of P. vivax sequences from the gorilla sample restricted the analysis to six genes and only five chimpanzee parasites. For these genes the gorilla P. vivax strain was quite divergent (Fig. 2B and SI Appendix, Fig. S3B), differing almost as much from the chimpanzee strains (on average 1.8%) as from the human strains (on average 2.4%). Whether the human P. vivax lineage falls within the radiation of the ape strains or groups as a sister clade depends on the position of the root of these trees. The closest available outgroup is P. cynomolgi, which is much more distant from the P. vivax sequences than they are from each other and may not root the tree reliably.
Fig. 2.
Evolutionary relationships of ape and human P. vivax strains. (A) An unrooted neighbor-joining tree constructed from a matrix of pairwise genetic distances from an alignment of 241 nuclear genes is shown for nine human (black) and six chimpanzee (red) P. vivax strains (the Inset shows the human P. vivax strains in greater detail). (B) As in A, but based on six nuclear genes with coverage in one gorilla P. vivax strain (green). The same human and chimpanzee P. vivax strains were included, except for PvSY81, which did not cover these genes. (C and D) Maximum-likelihood trees for fragments of nuclear genes PVP01_1418300 (C) and PVP01_1418500 (D) with P. cynomolgi and P. knowlesi included as outgroups. Sequences of P. carteri parasites are shown in blue. “Pv” denotes sequences from genome-wide analyses, shown in bold if generated by SWGA or derived from published data (SI Appendix, Table S2); all other sequences except for P. cynomolgi and P. knowlesi were generated by SGA and include a code identifying their geographic origin (SI Appendix, Fig. S5), ape subspecies (G.g.g., Gorilla gorilla gorilla; P.t.e., Pan troglodytes ellioti; P.t.t., Pan troglodytes troglodytes), and sample number (see SI Appendix, Table S4 for GenBank accession numbers). Bootstrap values ≥70 are shown for clades with two or more nonidentical tips. Fragment lengths in PvP01 are indicated above the trees. The scale bars indicate substitutions per site (see also SI Appendix, Fig. S3 for phylogenetic network representations).
To investigate further the relationships among ape and human P. vivax strains, we focused on the 10 genes that could be recovered from the single gorilla sample (SI Appendix, Table S2). Including both P. cynomolgi and P. knowlesi as outgroups, we found that four genes yielded a tree topology in which the human strains fell within the radiation of ape strains, while the six other genes yielded tree topologies in which human and ape parasites represented sister clades (SI Appendix, Fig. S4). To increase the number of geographically diverse ape P. vivax sequences, we used single genome amplification (SGA) to screen an existing collection of P. vivax-positive ape blood and fecal samples for five of these genes (SI Appendix, Fig. S5). Each DNA preparation was diluted so that only single DNA templates were amplified, which precludes in vitro recombination. For three of these genes, this analysis also yielded sequences from Plasmodium carteri, a rare parasite species thus far found in only two wild chimpanzees, which is distinct from but most closely related to the P. vivax clade (7, 30). When these additional sequences were included in the phylogenetic analyses, four of the five trees showed the human strains within the radiation of ape strains, including two in which the previous topology depicted ape and human strains as sister clades, with three of these four trees including gene sequences for P. carteri (Fig. 2C and SI Appendix, Fig. S6). Thus, the inclusion of gorilla P. vivax and/or P. carteri changed the topology from sister clades to a nested relationship. For the remaining gene, human and ape parasite sequences remained as sister clades, but only a single gorilla parasite sequence was available for analysis, and P. carteri sequences could not be amplified (Fig. 2D). These results indicate that the inferred relationships among P. vivax strains from apes and humans depend in large part on the number of available sequences, especially from gorilla parasites, as well as on the presence of a closely related outgroup.
Ape P. vivax Strains Maintain ORFs for rbp Genes That Are Pseudogenized in Human P. vivax.
Adaptation of Plasmodium parasites to new host species has been associated with gains and losses of genes encoding proteins involved in red blood cell invasion (13). We therefore compared the repertoire of P. vivax invasion genes in the genomes of human and chimpanzee parasites. Like human P. vivax, the chimpanzee parasite genomes contained genes encoding the Duffy-binding protein (DBP) and a related erythrocyte-binding protein (DBP2, or EBP) (31), but no additional DBP-like genes were identified.
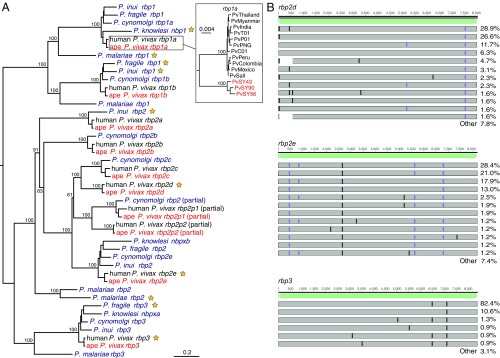
Variation in the complement of RBPs is thought to influence the ability of Plasmodium parasites to invade erythrocytes (13, 32). Human P. vivax, which exclusively invades reticulocytes, has full-length ORFs for five rbp genes (rbp1a, rbp1b, rbp2a, rbp2b, and rbp2c), all of which were conserved in the two chimpanzee P. vivax genomes PvSY56 and PvSY43. Two shorter rbp genes annotated in human P. vivax (rbp2p1 and rbp2p2) are believed to encode proteins that lack a C-terminal transmembrane domain; one of these, rbp2p1, appears to be present in all human P. vivax strains and also in P. cynomolgi, while rbp2p2 has been found only in a subset of human P. vivax strains (33). We identified orthologs of both of these partial genes in the chimpanzee P. vivax genomes (Fig. 3A), indicating that variation in the presence of rbp2p2 among human P. vivax strains is the result of a deletion after the divergence of human and ape parasites rather than a recent gene duplication. The finding of partial rbp2p1 and rbp2p2 genes in both ape and human P. vivax and of rbp2p1 in P. cynomolgi suggests that their encoded proteins have a conserved function. However, synonymous-to-nonsynonymous substitution (dN/dS)-based tests for positive selection (34) in invasion genes on the branch leading to human P. vivax (dbp, ebp, rbp1a, rbp1b, rbp2a, and rbp2b) failed to yield evidence of human-specific adaptation.
Fig. 3.
The rbp gene family in ape and human P. vivax. (A) A midpoint-rooted maximum-likelihood phylogenetic tree is shown depicting the relationships of human (black) and chimpanzee (PvSY56 and PvSY43, red) P. vivax rbp genes with their orthologs in P. knowlesi, P. cynomolgi, P. inui, P. fragile, and human P. malariae (purple). P. vivax, P. cynomolgi, and P. knowlesi genes are labeled according to their published names; genes from P. inui, P. fragile, and P. malariae are labeled according to the clade in which they are placed. Pseudogenes are indicated by yellow stars. The Inset shows the relationship of rbp1a sequences among representative human and three sequenced chimpanzee P. vivax strains, rooted using P. cynomolgi (see SI Appendix for details). (B) Locations of frameshift (purple) and premature stop (black) mutations in rbp2d, rbp2e, and rbp3 sequences assembled from published human P. vivax strains (20, 21), relative to the full-length coding sequence from chimpanzee P. vivax (light green). Each bar represents a set of mutations that occurred in two or more human P. vivax strains for which a full-length sequence was assembled (128, 162, and 227 sequences for rbp2d, rbp2e, and rbp3, respectively); the percentage of sequences containing the respective mutations is shown on the right, with “other” summarizing all mutations that occurred only once.
The human P. vivax genome also contains three rbp pseudogenes termed rbp2d, rbp2e, and rbp3. Seemingly functional orthologs of rbp2e and rbp3 are present in the genomes of the monkey parasites P. cynomolgi, P. knowlesi, and Plasmodium inui, while Plasmodium fragile has an intact rbp2e gene but a rbp3 pseudogene (Fig. 3A). So far, rbp2d has been identified only in P. vivax. The two chimpanzee P. vivax genomes PvSY56 and PvSY43 contained full-length intact ORFs corresponding to each of these three human P. vivax pseudogenes, indicating that the loss of function occurred after the divergence of human and ape parasites (Fig. 3A).
Since the pseudogenization of rbp2d, rbp2e, and rbp3 seems to be unique to the human lineage of P. vivax, we considered the possibility that these genes may be intact in some human strains. We mapped sequencing reads from 374 published human P. vivax strains (20, 21) to the rbp2d, rbp2e, and rbp3 reference genes and analyzed those that yielded a greater than threefold read coverage of the entire coding sequences. In each gene, we found at least one inactivating mutation that was present in all human parasite samples as well as numerous additional mutations that likely occurred subsequent to the initial pseudogenization event (Fig. 3B). The accumulation of additional frameshifts and stop mutations, some of which occur very close to the 5′ end of the coding sequence, suggests that these genes do not encode truncated proteins.
To examine whether the other chimpanzee P. vivax strains (SI Appendix, Table S1) contained any of the rbp2d-, rbp2e-, and rbp3-inactivating mutations, we mapped available sequencing reads to the respective PvSY56 genes. We also used SGA to amplify the same regions from P. vivax-positive gorilla samples. Although in most instances the coverage of the rbp2d, rbp2e, and rbp3 genes was incomplete, none of the recovered sequences contained the frameshift and stop codon mutations found in all human P. vivax strains (SI Appendix, Fig. S7). This was also true for SGA-derived gorilla parasite sequences from the multiply infected SAggg3157 sample, which contained a number of polymorphisms, none of which disrupted the respective reading frame. Thus, both chimpanzee and gorilla P. vivax parasites appear to maintain three genes encoding RBPs that have been lost in all human P. vivax strains, which could influence their host tropism.
Recombinant RBP2e and RBP3 Do Not Exhibit Species-Specific Red Blood Cell Binding.
The pseudogenization of rbp2d, rbp2e, and rbp3 in all human P. vivax strains raised the possibility that these proteins bind gorilla- and/or chimpanzee-specific erythrocyte receptors that are no longer used by the human parasite. Recombinant proteins comprising the N-terminal domain of RBPs encoded by human P. vivax (RBP2a160–1000 and RBP2b161–969) have been used to characterize their erythrocyte-binding properties (35, 36). These studies showed that RBP2b binds the reticulocyte-specific transferrin receptor 1 (TfR1), also termed “CD71” (35), while RBP2a binds an unknown receptor present on both normocytes and reticulocytes (36). To examine the function of chimpanzee P. vivax RBP2d, RBP2e, and RBP3 proteins, we expressed their N-terminal domains in bacteria for subsequent erythrocyte-binding studies. Although RBP2d165–967 could not be purified due to protein aggregation, RBP2e156–957 and RBP3149–968 were efficiently expressed and exhibited an α-helical and β-sheet content similar to human P. vivax RBP2b (SI Appendix, Fig. S8). Because some RBPs bind only reticulocytes, we attempted to enrich these cells from blood samples obtained from four humans, four chimpanzees, and one gorilla using a Percoll density gradient as previously described (35, 36). Despite repeated attempts, this approach yielded only partial reticulocyte enrichment for the ape blood samples, possibly due to differences in erythrocyte density between the different species (SI Appendix, SI Materials and Methods). Nonetheless, some enrichment of ape reticulocytes (up to 1.8%) was achieved as determined by thiazole orange (TO) staining.
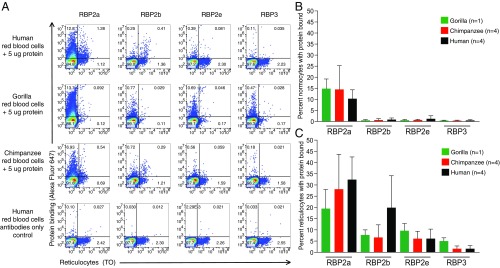
To examine binding to ape red blood cells, we first tested the two previously characterized human P. vivax RBP proteins, RBP2a160–1000 and RBP2b161–969 (35, 36). Ape and human red blood cells were incubated with each recombinant protein, and binding was assessed using protein-specific polyclonal rabbit antibodies followed by a fluorophore-labeled anti-rabbit antibody (35, 36). Reticulocytes were stained with TO before flow cytometry (Fig. 4A). Consistent with previous results, we observed robust binding of RBP2a to both human normocytes (10.3% of TO-negative cells) (Fig. 4B) and reticulocytes (32.4% of TO-positive cells) (Fig. 4C). Interestingly, a similar binding profile was observed for gorilla and chimpanzee red blood cells (Fig. 4 and SI Appendix, Fig. S9). As expected, RBP2b exhibited a strong preference for reticulocytes, binding 19.9% of human reticulocytes (Fig. 4C) but only a minor fraction (0.9%) of human normocytes (Fig. 4B), likely reflecting incomplete reticulocyte staining and/or nonspecific binding. RBP2b also bound chimpanzee and gorilla reticulocytes, albeit at a reduced level (Fig. 4C). Although the TfR1 proteins of chimpanzees and gorillas differ from their human counterpart by a few amino acids (SI Appendix, Fig. S10), none of these residues was identified as representing critical RBP2b contact sites (37). Thus, the decreased binding of RBP2b to ape reticulocytes is unlikely to be the result of sequence differences between chimpanzee, gorilla, and human TfR1 proteins and may instead reflect differences in TfR1 expression levels, posttranslational modifications, or other factors.
Fig. 4.
Binding of RBPs to ape and human red blood cells. (A) Dot plots depict the binding of human P. vivax RBP2a and RBP2b proteins and chimpanzee P. vivax RBP2e and RBP3 proteins to human (first row), gorilla (second row), and chimpanzee (third row) red blood cells, respectively, along with antibody-only controls of human red blood cells (fourth row). RBP binding was detected using an RBP-specific polyclonal rabbit antibody and an anti-rabbit (Alexa Fluor 647-labeled) secondary antibody (y axis), and reticulocytes were identified by staining with TO (x axis). Flow cytometry gates separating normocytes from reticulocytes and protein binding from no protein binding are shown by vertical and horizontal lines, respectively. Numbers indicate the percentage of total cells within the respective gate. (B) Percentage of gorilla (green), chimpanzee (red), and human (black) normocytes bound by the respective RBP. (C) Percentage of gorilla, chimpanzee, and human reticulocytes bound by the respective RBP. Experiments were performed as three technical replicates with the background signal from the antibody-only control subtracted from each binding result.
Having validated the experimental system, we next tested the binding of chimpanzee P. vivax RBP2e156–957 and RBP3149–968 to ape and human red blood cells. We found that neither of these two proteins bound particularly well to ape red blood cells, although RBP2e consistently yielded a higher signal than RBP3 (Fig. 4 B and C). Like the human P. vivax RBP2a and RBP2b proteins, RBP2e and RBP3 appeared to bind reticulocytes more efficiently than normocytes (Fig. 4 and SI Appendix, Fig. S9). However, there was no clear evidence for host specificity. Although RBP3 bound gorilla reticulocytes slightly more efficiently than chimpanzee and human reticulocytes, this result must be interpreted with caution, since only a single gorilla sample containing very few reticulocytes was available for testing (Fig. 4A). Indeed, when red blood cells from a macaque were tested, RBP2e and RBP3 were found to bind to reticulocytes from this host species also (SI Appendix, Fig. S9). To determine whether the low level of RBP2e and RBP3 binding was due to inefficient reticulocyte enrichment, we tested an additional chimpanzee blood sample with a particularly high reticulocyte count. Although Percoll gradient enrichment of this sample yielded twice as many reticulocytes (4%), this higher fraction did not improve RBP2e and RBP3 binding (SI Appendix, Fig. S11). Thus, the maintenance of the rbp2e and rbp3 genes in chimpanzee P. vivax cannot be readily explained by invoking interaction with a host-specific erythrocyte receptor.
Discussion
It has recently become apparent that wild-living African apes, including western and eastern gorillas as well as chimpanzees and bonobos, harbor malaria parasites that appear to be very closely related to P. vivax strains infecting humans in Asia and Central/South America (7, 8, 11). Since these results were based on a small number of mostly mitochondrial DNA fragments, we have now generated two nearly complete and several partial genome sequences from ape-infecting parasites. Analyses of these sequences show that ape and human parasites are indeed very closely related, with the human P. vivax sequences forming a monophyletic lineage either within or as a sister group to the ape parasites. The data also reveal that ape and human P. vivax strains exhibit distinct patterns of genetic diversity, reflecting very different demographic histories. The parasites infecting humans and apes are largely allopatric, but as yet there is no evidence that they represent distinct Plasmodium species. Thus, P. vivax in African apes represents a substantial and genetically diverse parasite reservoir from which future human infections could arise, even if eradication of current human strains were successful.
Recent papers have emphasized that, across the genome, the global genetic diversity of human P. vivax is somewhat greater than that of P. falciparum (21, 38). Here, we find that the level of neutral genetic diversity among P. vivax parasites from chimpanzees is about 10 times higher than that of human P. vivax strains. P. falciparum isolates also exhibit about 10 times less genetic diversity than is seen in closely related Laverania species (18, 19). This is consistent with both human parasites having undergone severe genetic bottlenecks. In the case of P. falciparum, this most likely occurred at the point when a gorilla parasite made the cross-species jump into humans (9, 30). For P. vivax, it is also possible that the ancestral parasites infected only nonhuman apes and that one of these crossed the species barrier and gave rise to the population of parasites currently infecting humans. Alternatively, ancient P. vivax may have infected all African ape species, including humans (7). In the latter case, the bottleneck would likely have occurred when P. vivax migrated with humans out of Africa, before the spread of the Duffy-negative mutation that eliminated P. vivax from humans in most (or at that time, perhaps all) of sub-Saharan Africa. While it is difficult to distinguish between these two scenarios on the basis of genetic data, we believe that the second scenario is the more likely. Ape P. vivax seems to circulate freely between chimpanzees and gorillas in the wild (7) and has caused disease in a Duffy-positive human (11). Consistent with this, the very high frequency of the Duffy-negative mutation suggests a long history of P. vivax exerting selective pressure on humans in Africa (39). It is likely that the geographic areas in which this mutation is most frequent (2) were influenced over a long timescale by the distribution of ape P. vivax to which humans were exposed.
The nature of the genetic diversity also differs markedly between the populations of P. vivax parasites that infect apes and humans. Among human strains, there is an unusually high fraction of nonsynonymous polymorphism. Moreover, these nonsynonymous polymorphisms exhibit a site-frequency spectrum that is almost identical to that seen for synonymous mutations, implying that they are segregating as if effectively neutral. Similar observations have been made for polymorphisms in P. falciparum (40), where this has been attributed, at least in part, to the repeated bottleneck events that the parasite undergoes in every life cycle at the obligate transmission events between host and vector, with rapid expansion in the human host (41). Here, we find that the ratio of nonsynonymous to synonymous mutations among the much more numerous polymorphisms among ape P. vivax strains is similar to the ratio at sites of divergence between P. vivax and its macaque relative P. cynomolgi. Thus, the unusual pattern of polymorphism in human P. vivax cannot reflect its life cycle but is more likely the consequence of the population having undergone a rapid expansion subsequent to the spread out of Africa.
The pseudogenization of three RBPs (rbp2d, rbp2e, and rbp3) in all human P. vivax strains could be taken to indicate human-specific adaptation after the parasite migrated out of Africa and no longer encountered chimpanzees or gorillas. However, our erythrocyte-binding results are not consistent with this scenario. We observed equivalent binding of RBP2e to ape and human red blood cells and only a modest increase in RBP3 binding to gorilla reticulocytes compared with human reticulocytes. Overall, the chimpanzee P. vivax-derived RBP2e and RBP3 proteins bound red blood cells much less well than the human P. vivax-derived RBP2a and RBP2b proteins, which could be due to low-affinity interactions, improper folding of the recombinant proteins, or the absence of other parasite proteins required for receptor engagement. We also considered the possibilities that RBP2e and RBP3 are not involved in erythrocyte binding or that the core receptor-binding domains were not included in the expressed proteins. However, others have shown that the P. knowlesi orthologs of RBP3 and RBP2e, termed “PkNBPXa” and “PkNBPXb,” both bind red blood cells (42) and that the binding domain of PkNBPXb maps to a region included in our RBP2e construct (42). Moreover, deletion of PkNBPXa severely reduced the ability of P. knowlesi merozoites to invade human red blood cells in vitro (43), even though all human-infecting P. vivax strains have lost RBP3. It could be argued that the large number of RBP genes in P. vivax (eight full-length and two partial genes) compared with P. knowlesi (two full-length genes and one pseudogene) provides functional redundancy that compensates for the loss of rbp genes in human P. vivax. However, this would also apply to ape P. vivax, where pseudogenes have not been found. Instead, the loss of rbp genes may be slightly deleterious and incur a fitness cost. In ape P. vivax, such an inactivating mutation would likely be selected against and thus not spread in the population. However, in human P. vivax the same mutation could be effectively neutral, as many nonsynonymous mutations appear to be, and so it could survive and drift to fixation. Whether this explains the loss of rbp2d, rbp2e, and rbp3 in human P. vivax remains unknown. However, we found no evidence that rbp2e and rbp3 genes are maintained in ape P. vivax parasites because they interact with chimpanzee- and/or gorilla-specific erythrocyte receptors that are absent from human red blood cells.
Despite their common origin in Africa, ape and human P. vivax populations have likely had little or no geographic overlap subsequent to the escape of the human-infecting lineage out of Africa. Under the bottleneck scenario, the most recent common ancestor of human P. vivax was in the lineage that emerged from Africa. This may have been coincident with the emergence of modern humans from Africa, perhaps around 75,000 y ago (44). Molecular clock estimates have placed the P. vivax most recent common ancestor (MRCA) at least 50,000–70,000 y ago (6, 45), but these relied on rate assumptions that may not be accurate. We have argued that the MRCA of P. falciparum strains may have existed within the last 10,000 y (18) despite molecular clock estimates that place the origin of that species much earlier. Similarly, the MRCA of human P. vivax may have left Africa in a more recent wave of human migration, although its higher levels of genetic diversity (21) suggest that human P. vivax is older than P. falciparum. Once out of Africa, P. vivax spread through Asia and Europe and probably from Europe into the Americas (39, 46). Strains of P. vivax now present in Madagascar and East Africa, in areas where nonhuman apes are absent, likely reflect reintroductions from Asia (47). Given this historical isolation of ape and human P. vivax strains, substantial gene flow between the two populations is unlikely. The mixture of topologies found for different genes, with some showing separation of the ape and human parasite lineages and others having the human parasites nested within the radiation of the ape parasites, likely reflects an ongoing process of lineage sorting in the absence of introgression. However, this does not mean that the two populations have become isolated species. Both ape and human P. vivax exhibit broad natural tropism (7, 11, 15, 30), and it therefore seems very likely that ape and human strains could infect the same hosts and undergo genetic exchange if their geography overlapped. In recent years, reports of P. vivax infection of African humans have been increasing, including instances of infection of Duffy-negative individuals (4). It will be important to monitor these cases to determine whether any reflect zoonotic transmissions from apes and whether there is any sign of introgression between ape- and human-infecting strains.
Methods
Sample Collection, DNA Extraction, and Plasmodium Screening.
Blood samples were obtained from chimpanzees at the Sanaga-Yong Chimpanzee Rescue Center following routine veterinary examination. Blood was also collected from a wild-living habituated chimpanzee from the Tai Forest in Côte d’Ivoire during emergency surgery (48). The gorilla blood sample (Gor3157, also referred to as “SAggg3157”) was obtained from confiscated bushmeat in Cameroon. All samples were shipped in compliance with Convention on International Trade in Endangered Species of Wild Fauna and Flora regulations and country-specific import and export permits. DNA was extracted from whole-blood samples using the QIAmp Blood DNA Mini Kit or the Puregene Core Blood Kit (Qiagen) and was screened for Plasmodium using both pan-Plasmodium primers and P. vivax-specific primers, as described (7, 9). Amplicons were sequenced using Sanger or MiSeq technologies (SI Appendix).
SWGA.
Nearly full-length and partial P. vivax genomes were amplified from unprocessed chimpanzee and gorilla blood as described (18, 23) using multiple rounds of SWGA with different primer sets, with and without prior digestion with MspJI and FspEI to selectively degrade host DNA (SI Appendix, SI Materials and Methods and Table S1).
Illumina and PacBio Sequencing.
Short insert libraries were prepared from pooled SWGA products of samples SY43 and SY56 using a KAPA HyperPlus Kit and were MiSeq sequenced. Pooled SWGA products were also linearized with S1 nuclease (Promega), needle-sheared to reduce fragment size, and subjected to PacBio SMRT Cell sequencing (University of Delaware Sequencing Core). SWGA products from samples SY81, SY90, Sagu, and Gor3157 were Illumina sequenced to obtain partial P. vivax genomes (SI Appendix).
Assembly of Chimpanzee P. vivax Draft Genomes.
Illumina sequencing reads were error-corrected using SPAdes (49) and were mapped to the chimpanzee reference genome; unmapped reads were mapped to the P. vivax P01 reference genome (24). Regions with low coverage or poor paired-read support were removed. Gaps were closed using FGAP (50) with proovread-corrected PacBio reads (51) followed by iterations of GapFiller (52) and IMAGE (53). The PvSY56 draft genome was further improved by de novo assembly of subtelomeric reads using SPAdes (49). Annotations were transferred to the final assembly using RATT (54) and were predicted using Companion (55) in the de novo-assembled subtelomeres, followed by manual correction. Genome annotations for PvSY43 and PvSY56 are available upon request. Sets of orthologous genes from PvSY43, PvSY56, PvP01, and PvSalI were masked using segmasker (nebc.nox.ac.uk/bioinformatics/docs/blast+.html) to exclude low-complexity regions and were aligned using TranslatorX/MUSCLE (56). Outgroup sequences, where available, were added to these alignments using MUSCLE. Divergence between sequences was calculated in R using ape (57) with no correction for multiple substitutions (raw model). Fourfold and zerofold degenerate sites were extracted if the classification was true for all sequences in the alignment (SI Appendix).
SNP Calling.
In addition to sequencing SWGA products, we also mined publicly available databases for human (21) and chimpanzee (14) P. vivax reads. SNPs were called following best practices for the Genome Analysis Toolkit with hard filtering (58), using PvSY56 and PvP01 as ape and human P. vivax reference sequences, respectively. Regions classified as low-complexity by dustmasker, genes in subtelomeric and internal hypervariable regions (20), and all vir and phist genes were excluded. Sites with coverage of at least five reads were considered callable, with genes containing fewer than 60% of the sites of the reference excluded. Only sites callable for all strains were analyzed. Site-frequency spectra were generated from high-quality SNP calls for all high-coverage Southeast Asian P. vivax strains in the MalariaGEN Genome Variation project (20). Fourfold and zerofold degenerate sites were identified in the PvSalI reference sequence using custom R scripts, and derived alleles at polymorphic sites were identified using the est-sfs unfolder (59) with PvSY56 and P. cynomolgi as outgroups (SI Appendix).
SGA of Geographically Diverse P. vivax Strains.
Ape P. vivax and P. carteri sequences were amplified as described from stored ape blood and fecal samples previously shown to be positive for these parasites (7). Fragments of five genes were targeted using newly designed primers (SI Appendix).
Identification of rbp Genes.
Chimpanzee P. vivax rbp genes were identified during annotation of the PvSY56 and PvSY43 genomes. rbp orthologs from P. malariae strain PmUG01 (14), P. knowlesi strain H (ref. 60, 2015 update), P. cynomolgi strains B and Berok (61, 62), P. inui strain San Antonio 1, and P. fragile strain Nilgiri (PlasmoDB) were identified from annotations and using P. vivax rbp genes as query sequences. Human P. vivax sequencing reads (20, 21) were downloaded and mapped to rbp2e (PVP01_0700500), rbp2p1 (PVP01_0534400), rbp2p2 (PVP01_101590), rbp3 (PVP01_1469400), and rbp2d (PVP01_1471400 and PVX_101585), with a consensus sequence called for all positions with threefold or greater coverage. The positions of frameshifts and stop codons were identified relative to the ape P. vivax sequence (SI Appendix).
RBP Expression and Red Blood Cell-Binding Assays.
Chimpanzee P. vivax rbp2d, rbp2e, and rbp3 sequences were codon optimized, synthesized, and cloned into pET-32a(+) (Novagen), yielding constructs RBP2d165–967, RBP2e156–957, and RBP3149–968, respectively. Proteins were expressed using Escherichia coli SHuffle T7 (New England Biolabs) and purified using a HisTrap (GE Healthcare) column. After hexahistidine tag cleavage and purification through a Q-Sepharose HiTrap column (GE Healthcare), fractions containing the respective proteins (determined by SDS PAGE) were concentrated and further purified by size-exclusion chromatography. RBP2d165–967 could not be purified due to protein aggregation. Expression and purification of two human P. vivax RBP proteins, RBP2a160–1000 and RBP2b161–969, were performed as described (35, 36).
Whole blood from five chimpanzees (New Iberia Research Center), one gorilla (Lincoln Park Zoo), and one rhesus macaque (BioIVT) was collected in ACD-A collection tubes (BD Biosciences). All ape blood samples were leftover specimens obtained during routine health screenings (the macaque blood was purchased). Blood was also obtained from healthy human volunteers at the University of Pennsylvania under Internal Review Board protocol 813699. White blood cells were first removed by leukocyte filtration, and reticulocytes were subsequently enriched by spinning red blood cells (50% hematocrit) through a 65–75% (vol/vol) isotonic Percoll cushion and collecting the cell band at the interface. To assess RBP binding, recombinant protein was incubated with red blood cells for 1 h, followed by detection with an RBP-specific polyclonal rabbit antibody and an anti-rabbit (Alexa Fluor 647-labeled) secondary antibody (Thermo Fisher Scientific). Between each incubation step, cells were washed in 180 μL PBS containing 1% BSA (Sigma). Cells were stained in the dark with BD Retic-Count reagent for 30 min at room temperature, spun, and resuspended in 1.2 mL of PBS before analysis on an Accuri flow cytometer (BD). Experiments were performed as three technical replicates with the background signal from the antibody-only control subtracted from each binding result (SI Appendix).
Supplementary Material
Acknowledgments
We thank Andrew Smith, Catherine Bahari, and Alex Shazad for performing Illumina sequencing; the University of Delaware Sequencing Core for PacBio sequencing; Dana Bellissimo for assistance with flow cytometry; the staff of the Sanaga-Yong Rescue Center and Jane Fontenot, Melany Musso, and Francois Villinger at the New Iberia Research Center for providing leftover blood samples from captive chimpanzees; Marisa Shender from the Lincoln Park Zoo for providing leftover gorilla blood; the Taї Chimpanzee Project assistants for field work; the Cameroonian Ministries of Health, Forestry and Wildlife, and Scientific Research and Innovation for permission to perform studies in Cameroon; and the Ministries of Environment and Forests and Research, Office Ivoirien des Parcs et Réserves, Taï National Park, and the Swiss Centre for Scientific Research for permission to perform studies in Côte d’Ivoire. This work was supported in part by NIH Grants R01 AI097137, R01 AI091595, R37 AI050529, and P30 AI045008 (to B.H.H.); a grant from the Agence Nationale de Recherche (Programme Blanc, Sciences de la Vie, de la Santé et des Ecosystémes, ANR 11 BSV3 021 01, Projet PRIMAL) (to M.P.); and an Australian Research Council Future Fellowship and Howard Hughes Medical Institute–Wellcome Trust International Research Scholar Award 208693/Z/17/Z (to W.-H.T.). D.E.L. was supported by an NIH Training Grant T32 AI 007532; O.A.M. was supported by Biotechnology and Biological Sciences Research Council Grant BB/M010996/1 (EASTBIO); and A.L.K.M. was supported by Wellcome Trust PhD Programme Grant 108905/Z/15/Z.
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the GenBank database (accession nos. PRJNA474492 and MH443154–MH443228).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1810053115/-/DCSupplemental.
References
- 1.World Health Organisation 2017 World malaria report 2017. Available at www.who.int/malaria/publications/world-malaria-report-2017/report/en/. Accessed March 1, 2018.
- 2.Howes RE, et al. The global distribution of the Duffy blood group. Nat Commun. 2011;2:266. doi: 10.1038/ncomms1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miller LH, Mason SJ, Clyde DF, McGinniss MH. The resistance factor to Plasmodium vivax in blacks. The Duffy-blood-group genotype, FyFy. N Engl J Med. 1976;295:302–304. doi: 10.1056/NEJM197608052950602. [DOI] [PubMed] [Google Scholar]
- 4.Zimmerman PA. Plasmodium vivax infection in Duffy-negative people in Africa. Am J Trop Med Hyg. 2017;97:636–638. doi: 10.4269/ajtmh.17-0461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cornejo OE, Escalante AA. The origin and age of Plasmodium vivax. Trends Parasitol. 2006;22:558–563. doi: 10.1016/j.pt.2006.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mu J, et al. Host switch leads to emergence of Plasmodium vivax malaria in humans. Mol Biol Evol. 2005;22:1686–1693. doi: 10.1093/molbev/msi160. [DOI] [PubMed] [Google Scholar]
- 7.Liu W, et al. African origin of the malaria parasite Plasmodium vivax. Nat Commun. 2014;5:3346. doi: 10.1038/ncomms4346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu W, et al. Wild bonobos host geographically restricted malaria parasites including a putative new Laverania species. Nat Commun. 2017;8:1635. doi: 10.1038/s41467-017-01798-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu W, et al. Origin of the human malaria parasite Plasmodium falciparum in gorillas. Nature. 2010;467:420–425. doi: 10.1038/nature09442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaiser M, et al. Wild chimpanzees infected with 5 Plasmodium species. Emerg Infect Dis. 2010;16:1956–1959. doi: 10.3201/eid1612.100424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Prugnolle F, et al. Diversity, host switching and evolution of Plasmodium vivax infecting African great apes. Proc Natl Acad Sci USA. 2013;110:8123–8128. doi: 10.1073/pnas.1306004110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Faust C, Dobson AP. Primate malarias: Diversity, distribution and insights for zoonotic Plasmodium. One Health. 2015;1:66–75. doi: 10.1016/j.onehlt.2015.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scully EJ, Kanjee U, Duraisingh MT. Molecular interactions governing host-specificity of blood stage malaria parasites. Curr Opin Microbiol. 2017;40:21–31. doi: 10.1016/j.mib.2017.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rutledge GG, et al. Plasmodium malariae and P. ovale genomes provide insights into malaria parasite evolution. Nature. 2017;542:101–104. doi: 10.1038/nature21038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buery JC, et al. Mitochondrial genome of Plasmodium vivax/simium detected in an endemic region for malaria in the Atlantic forest of Espírito Santo state, Brazil: Do mosquitoes, simians and humans harbour the same parasite? Malar J. 2017;16:437. doi: 10.1186/s12936-017-2080-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brasil P, et al. Outbreak of human malaria caused by Plasmodium simium in the Atlantic forest in Rio de Janeiro: A molecular epidemiological investigation. Lancet Glob Health. 2017;5:e1038–e1046. doi: 10.1016/S2214-109X(17)30333-9. [DOI] [PubMed] [Google Scholar]
- 17.Brock PM, et al. Plasmodium knowlesi transmission: Integrating quantitative approaches from epidemiology and ecology to understand malaria as a zoonosis. Parasitology. 2016;143:389–400. doi: 10.1017/S0031182015001821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sundararaman SA, et al. Genomes of cryptic chimpanzee Plasmodium species reveal key evolutionary events leading to human malaria. Nat Commun. 2016;7:11078. doi: 10.1038/ncomms11078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Otto TD, et al. Genomes of all known members of a Plasmodium subgenus reveal paths to virulent human malaria. Nat Microbiol. 2018;3:687–697. doi: 10.1038/s41564-018-0162-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pearson RD, et al. Genomic analysis of local variation and recent evolution in Plasmodium vivax. Nat Genet. 2016;48:959–964. doi: 10.1038/ng.3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hupalo DN, et al. Population genomics studies identify signatures of global dispersal and drug resistance in Plasmodium vivax. Nat Genet. 2016;48:953–958. doi: 10.1038/ng.3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leichty AR, Brisson D. Selective whole genome amplification for resequencing target microbial species from complex natural samples. Genetics. 2014;198:473–481. doi: 10.1534/genetics.114.165498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cowell AN, et al. Selective whole-genome amplification is a robust method that enables scalable whole-genome sequencing of Plasmodium vivax from unprocessed clinical samples. MBio. 2017;8:e02257. doi: 10.1128/mBio.02257-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Auburn S, et al. A new Plasmodium vivax reference sequence with improved assembly of the subtelomeres reveals an abundance of pir genes. Wellcome Open Res. 2016;1:4. doi: 10.12688/wellcomeopenres.9876.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rand DM, Kann LM. Excess amino acid polymorphism in mitochondrial DNA: Contrasts among genes from Drosophila, mice, and humans. Mol Biol Evol. 1996;13:735–748. doi: 10.1093/oxfordjournals.molbev.a025634. [DOI] [PubMed] [Google Scholar]
- 26.McDonald JH, Kreitman M. Adaptive protein evolution at the Adh locus in Drosophila. Nature. 1991;351:652–654. doi: 10.1038/351652a0. [DOI] [PubMed] [Google Scholar]
- 27.Chuquiyauri R, et al. Genome-scale protein microarray comparison of human antibody responses in Plasmodium vivax relapse and reinfection. Am J Trop Med Hyg. 2015;93:801–809. doi: 10.4269/ajtmh.15-0232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang B, et al. Immunoprofiling of the tryptophan-rich antigen family in Plasmodium vivax. Infect Immun. 2015;83:3083–3095. doi: 10.1128/IAI.03067-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Keightley PD, Campos JL, Booker TR, Charlesworth B. Inferring the frequency spectrum of derived variants to quantify adaptive molecular evolution in protein-coding genes of Drosophila melanogaster. Genetics. 2016;203:975–984. doi: 10.1534/genetics.116.188102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Loy DE, et al. Out of Africa: Origins and evolution of the human malaria parasites Plasmodium falciparum and Plasmodium vivax. Int J Parasitol. 2017;47:87–97. doi: 10.1016/j.ijpara.2016.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hester J, et al. De novo assembly of a field isolate genome reveals novel Plasmodium vivax erythrocyte invasion genes. PLoS Negl Trop Dis. 2013;7:e2569. doi: 10.1371/journal.pntd.0002569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cowman AF, Tonkin CJ, Tham WH, Duraisingh MT. The molecular basis of erythrocyte invasion by malaria parasites. Cell Host Microbe. 2017;22:232–245. doi: 10.1016/j.chom.2017.07.003. [DOI] [PubMed] [Google Scholar]
- 33.Hietanen J, et al. Gene models, expression repertoire, and immune response of Plasmodium vivax reticulocyte binding proteins. Infect Immun. 2015;84:677–685. doi: 10.1128/IAI.01117-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 35.Gruszczyk J, et al. Transferrin receptor 1 is a reticulocyte-specific receptor for Plasmodium vivax. Science. 2018;359:48–55. doi: 10.1126/science.aan1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gruszczyk J, et al. Structurally conserved erythrocyte-binding domain in Plasmodium provides a versatile scaffold for alternate receptor engagement. Proc Natl Acad Sci USA. 2016;113:E191–E200. doi: 10.1073/pnas.1516512113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gruszczyk J, et al. Cryo-EM structure of an essential Plasmodium vivax invasion complex. Nature. 2018;559:135–139. doi: 10.1038/s41586-018-0249-1. [DOI] [PubMed] [Google Scholar]
- 38.Neafsey DE, et al. The malaria parasite Plasmodium vivax exhibits greater genetic diversity than Plasmodium falciparum. Nat Genet. 2012;44:1046–1050. doi: 10.1038/ng.2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carter R. Speculations on the origins of Plasmodium vivax malaria. Trends Parasitol. 2003;19:214–219. doi: 10.1016/s1471-4922(03)00070-9. [DOI] [PubMed] [Google Scholar]
- 40.Chang H-H, et al. Genomic sequencing of Plasmodium falciparum malaria parasites from Senegal reveals the demographic history of the population. Mol Biol Evol. 2012;29:3427–3439. doi: 10.1093/molbev/mss161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chang H-H, et al. Malaria life cycle intensifies both natural selection and random genetic drift. Proc Natl Acad Sci USA. 2013;110:20129–20134. doi: 10.1073/pnas.1319857110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Semenya AA, Tran TM, Meyer EV, Barnwell JW, Galinski MR. Two functional reticulocyte binding-like (RBL) invasion ligands of zoonotic Plasmodium knowlesi exhibit differential adhesion to monkey and human erythrocytes. Malar J. 2012;11:228. doi: 10.1186/1475-2875-11-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moon RW, et al. Normocyte-binding protein required for human erythrocyte invasion by the zoonotic malaria parasite Plasmodium knowlesi. Proc Natl Acad Sci USA. 2016;113:7231–7236. doi: 10.1073/pnas.1522469113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pagani L, et al. Genomic analyses inform on migration events during the peopling of Eurasia. Nature. 2016;538:238–242. doi: 10.1038/nature19792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McManus KF, et al. Population genetic analysis of the DARC locus (Duffy) reveals adaptation from standing variation associated with malaria resistance in humans. PLoS Genet. 2017;13:e1006560. doi: 10.1371/journal.pgen.1006560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gelabert P, et al. Mitochondrial DNA from the eradicated European Plasmodium vivax and P. falciparum from 70-year-old slides from the Ebro Delta in Spain. Proc Natl Acad Sci USA. 2016;113:11495–11500. doi: 10.1073/pnas.1611017113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Culleton R, et al. The origins of African Plasmodium vivax; insights from mitochondrial genome sequencing. PLoS One. 2011;6:e29137. doi: 10.1371/journal.pone.0029137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Köndgen S, et al. Pasteurella multocida involved in respiratory disease of wild chimpanzees. PLoS One. 2011;6:e24236. doi: 10.1371/journal.pone.0024236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bankevich A, et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Piro VC, et al. FGAP: An automated gap closing tool. BMC Res Notes. 2014;7:371. doi: 10.1186/1756-0500-7-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hackl T, Hedrich R, Schultz J, Förster F. proovread: Large-scale high-accuracy PacBio correction through iterative short read consensus. Bioinformatics. 2014;30:3004–3011. doi: 10.1093/bioinformatics/btu392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nadalin F, Vezzi F, Policriti A. GapFiller: A de novo assembly approach to fill the gap within paired reads. BMC Bioinformatics. 2012;13(Suppl 14):S8. doi: 10.1186/1471-2105-13-S14-S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tsai IJ, Otto TD, Berriman M. Improving draft assemblies by iterative mapping and assembly of short reads to eliminate gaps. Genome Biol. 2010;11:R41. doi: 10.1186/gb-2010-11-4-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Otto TD, Dillon GP, Degrave WS, Berriman M. RATT: Rapid annotation transfer tool. Nucleic Acids Res. 2011;39:e57. doi: 10.1093/nar/gkq1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Steinbiss S, et al. Companion: A web server for annotation and analysis of parasite genomes. Nucleic Acids Res. 2016;44:W29–W34. doi: 10.1093/nar/gkw292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Abascal F, Zardoya R, Telford MJ. TranslatorX: Multiple alignment of nucleotide sequences guided by amino acid translations. Nucleic Acids Res. 2010;38:W7–W13. doi: 10.1093/nar/gkq291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Paradis E, Claude J, Strimmer K. APE: Analyses of phylogenetics and evolution in R language. Bioinformatics. 2004;20:289–290. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- 58.Van der Auwera GA, et al. From FastQ data to high confidence variant calls: The genome analysis toolkit best practices pipeline. Curr Protoc Bioinformatics. 2013;43:11.10.1–11.10.33. doi: 10.1002/0471250953.bi1110s43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Keightley PD, Jackson BC. Inferring the probability of the derived vs. the ancestral allelic state at a polymorphic site. Genetics. 2018;209:897–906. doi: 10.1534/genetics.118.301120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pain A, et al. The genome of the simian and human malaria parasite Plasmodium knowlesi. Nature. 2008;455:799–803. doi: 10.1038/nature07306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pasini EM, et al. An improved Plasmodium cynomolgi genome assembly reveals an unexpected methyltransferase gene expansion. Wellcome Open Res. 2017;2:42. doi: 10.12688/wellcomeopenres.11864.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tachibana S, et al. Plasmodium cynomolgi genome sequences provide insight into Plasmodium vivax and the monkey malaria clade. Nat Genet. 2012;44:1051–1055. doi: 10.1038/ng.2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.