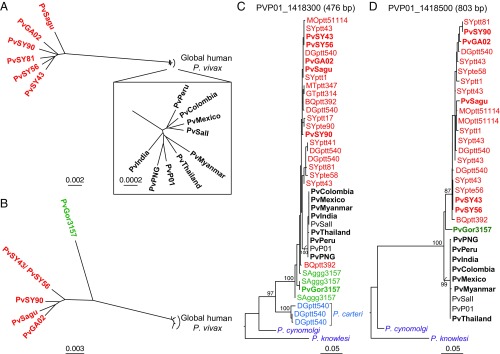
Fig. 2.
Evolutionary relationships of ape and human P. vivax strains. (A) An unrooted neighbor-joining tree constructed from a matrix of pairwise genetic distances from an alignment of 241 nuclear genes is shown for nine human (black) and six chimpanzee (red) P. vivax strains (the Inset shows the human P. vivax strains in greater detail). (B) As in A, but based on six nuclear genes with coverage in one gorilla P. vivax strain (green). The same human and chimpanzee P. vivax strains were included, except for PvSY81, which did not cover these genes. (C and D) Maximum-likelihood trees for fragments of nuclear genes PVP01_1418300 (C) and PVP01_1418500 (D) with P. cynomolgi and P. knowlesi included as outgroups. Sequences of P. carteri parasites are shown in blue. “Pv” denotes sequences from genome-wide analyses, shown in bold if generated by SWGA or derived from published data (SI Appendix, Table S2); all other sequences except for P. cynomolgi and P. knowlesi were generated by SGA and include a code identifying their geographic origin (SI Appendix, Fig. S5), ape subspecies (G.g.g., Gorilla gorilla gorilla; P.t.e., Pan troglodytes ellioti; P.t.t., Pan troglodytes troglodytes), and sample number (see SI Appendix, Table S4 for GenBank accession numbers). Bootstrap values ≥70 are shown for clades with two or more nonidentical tips. Fragment lengths in PvP01 are indicated above the trees. The scale bars indicate substitutions per site (see also SI Appendix, Fig. S3 for phylogenetic network representations).