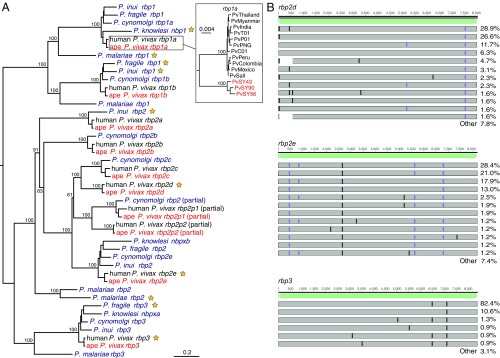
Fig. 3.
The rbp gene family in ape and human P. vivax. (A) A midpoint-rooted maximum-likelihood phylogenetic tree is shown depicting the relationships of human (black) and chimpanzee (PvSY56 and PvSY43, red) P. vivax rbp genes with their orthologs in P. knowlesi, P. cynomolgi, P. inui, P. fragile, and human P. malariae (purple). P. vivax, P. cynomolgi, and P. knowlesi genes are labeled according to their published names; genes from P. inui, P. fragile, and P. malariae are labeled according to the clade in which they are placed. Pseudogenes are indicated by yellow stars. The Inset shows the relationship of rbp1a sequences among representative human and three sequenced chimpanzee P. vivax strains, rooted using P. cynomolgi (see SI Appendix for details). (B) Locations of frameshift (purple) and premature stop (black) mutations in rbp2d, rbp2e, and rbp3 sequences assembled from published human P. vivax strains (20, 21), relative to the full-length coding sequence from chimpanzee P. vivax (light green). Each bar represents a set of mutations that occurred in two or more human P. vivax strains for which a full-length sequence was assembled (128, 162, and 227 sequences for rbp2d, rbp2e, and rbp3, respectively); the percentage of sequences containing the respective mutations is shown on the right, with “other” summarizing all mutations that occurred only once.