Abstract
The activating transcription factor (ATF) 4 belongs to the ATF/CREB (cAMP Response Element Binding bZIP [Basic Leucine Zipper]) transcription factor family, and plays a central role in the UPR (Unfolded Protein Response) process in cells. The induction of ATF4 expression has previously been shown to increase the replication of HIV-1. However, the detailed mechanism underlying this effect and the factors involved in the regulation of ATF4 function are still unknown. Here, we demonstrate first that knocking out ATF4 using siRNA shows a strong negative effect on HIV-1 production, indicating that ATF4 is a functional positive cellular factor in HIV-1 production. To determine the mechanism by which ATF4 regulates the HIV-1 life cycle, we assessed the effect of the overexpression of wild type ATF4 and its various derivatives on HIV-1 LTR-mediated transcriptional activation and the production of HIV-1 particles. This effect was studied through co-transfection experiments with either reporter vectors or proviral DNA. We found that the N-terminal domains of ATF4 are involved in HIV-1 LTR-mediated transcriptional activation, and thus in HIV-1 production.
Keywords: Activating Transctiption Factor 4, ATF4, HIV-1, HIV-1 LTR, HIV-1 promoter, Transctiption activation
INTRODUCTION
The activating transcription factor (ATF) 4, which is composed of 351 amino acids, is a member of the ATF/CREB (cAMP Response Element Binding) protein family. It could function interestingly as either a transcriptional activator or a repressor (1, 2). It is also known to act as a protective factor that mediates the adaptation of cells to stressors such as endoplasmic reticulum (ER) stress and nutrient deprivation (3). These stressors induce ATF4 expression, resulting in the activation of the unfolded protein response (UPR) pathway, which in turn increases the translation of ATF4 but reduces global protein synthesis (1, 4, 5). ATF4 is capable of forming both homodimers and heterodimers (6). The N-Terminus of ATF4, which is called the p300 interaction site, functions as a transcriptional activation domain and is implicated in protein stability as well (7–9). A basic-leucine zipper (bZIP) domain is located in the C-Terminus of ATF4 and is known to interact with DNA (6, 10). The leucine zipper of the bZIP domain contains alpha helices with leucine residues that mediate dimerization with a parallel leucine zipper domain (11). ATF4 can be localized to the nucleus by a nuclear localization signal sequence KKLKK (amino acids 280 to 284) in the basic region (12). The expression of ATF4 could be stimulated or blocked by viral infection, causing either an increase or decrease in viral production (13–21). Infection with the human cytomegalovirus (HCMV) activates the UPR pathway, and the HCMV proteins activate ATF4 expression (13, 14). The transcriptional activity and thus the production of the murine cytomegalovirus (MCMV) are known to be mediated by the binding of ATF4 to DNA and the subsequent activation of transcription (15). Dengue virus (DENV) can also increase the expression of ATF4 in the nucleus (16). The infectious bronchitis virus (IBV), which is a coronavirus, induces ATF4 expression at early stages of infection (17). It is also known that ATF4 expression is induced by the human herpesvirus 8 (HHV-8), resulting in the upregulation of HHV-8 production (18). On the other hand, several studies have shown that the expression of ATF4 could be blocked by certain types of viruses, such as myxoma virus (MXMV) (19), chikungunya virus (CHIKV) (20), and herpes simplex virus-1 (HSV-1) (21).
In acute infections with HIV, ATF4 accumulates (22). ATF/CREB proteins can bind to a region on the HIV-1 LTR (23, 24) and thereby increase HIV-1 LTR activation and replication synergistically with the viral protein Tat (22). In addition, ATF4 interacts with the Tax protein of the human T cell leukemia virus (HTLV-1) and regulates LTR activation (25). Thus, ATF4 appears to have a positive role in regulating HIV-1, but the exact molecular mechanism of ATF4-mediated HIV-1 activation still remains to be determined. To address this knowledge gap, we examined the effect of the overexpression of wild type ATF4 and its various derivatives on the production of HIV-1 through transfection experiments of HIV-1 proviral DNA. We have also determined through ATF4 knock-down study that ATF4 is a functional positive factor in HIV-1 production.
RESULTS AND DISCUSSION
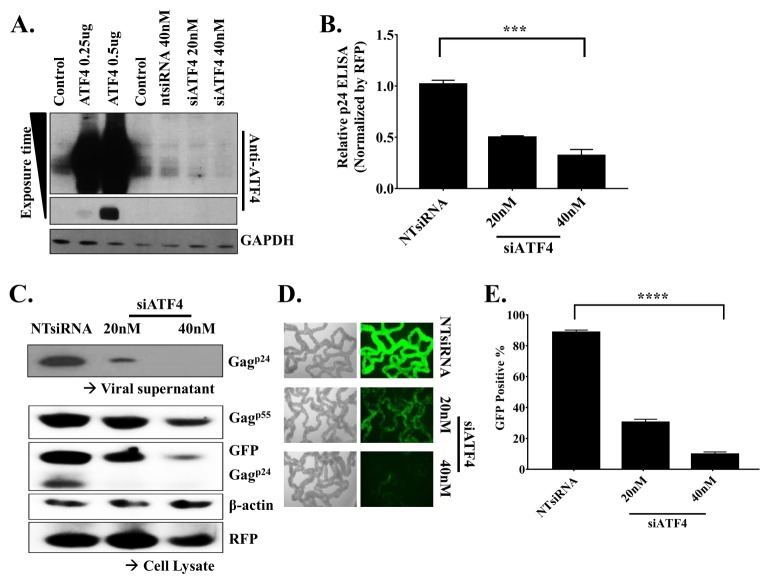
ATF4 is functionally required in HIV-1 production
To examine first whether ATF4 is required in HIV-1 production, we transfected siATF4 into 293T cells in varying doses. Lysates from the transfected cells were subjected to SDS-PAGE and immunoblot analysis using anti-ATF4 and anti-GAPDH antibodies as probes. ATF4, with a molecular weight of 50 kDa, was determined. Based on the observed band intensities, ATF4 expression was found to decrease with increasing concentrations of siATF4 (Fig. 1A). Next, we knocked down ATF4 expression using siRNA to investigate the effect of ATF4 on HIV-1 replication and infection. We transiently transfected 293T cells with 250 ng of pNL4-3/GFP, 100 ng of the RFP expression plasmid (pDs-red expression N1, internal control), and varying concentrations of siATF4. Virus production was assessed by measuring the amount of HIV-1 p24 antigen in constant volumes of viral supernatants from the transfected cells through an enzyme-linked immunosorbent assay (ELISA). The relative production of p24 decreased with increasing concentrations of siATF4 (Fig. 1B). To confirm the effect of siATF4 on HIV-1 production, cell lysates and viral supernatants were subjected to SDS-PAGE and immunoblot analysis using anti-p24, anti-GFP, anti-β-actin, and anti-RFP antibodies as probes (Fig. 1C). The intensity of the band corresponding to the viral p24 decreased with increasing concentrations of siATF4. Additionally, the intensities of the bands corresponding to p55, GFP, and p24 were consistent with those in previous data. In order to further characterize the effect of siATF4 on virus production, we re-infected fresh MT-4 cells with constant volumes of viral supernatant from 293T cells with knocked down ATF4 expression. Using an inverted fluorescent microscope, we observed that the GFP signal in MT-4 cells decreased with increasing concentrations of siATF4 (Fig. 1D). We then quantified the cells using FACS. The number of GFP-positive cells was dramatically reduced when the concentration of siATF4 was increased (Fig. 1E). Our data showed that ATF4 strongly inhibits HIV-1 production. These results suggest that ATF4 is necessary in the regulation of the HIV-1 life cycle.
Fig. 1.
ATF4 is functionally required in HIV-1 production. (A) Western blot results of overexpression and Knockdown of ATF4. (B) Virus titer of viral soup was determined by p24Gag ELISA then normalized by RFP level, % of Non-targeting siRNA. (C) Western blot results of viral supernatant and cell lysate. (D, E) MT-4 cells were infected with the same volume of viral supernatant generated from ATF4 knock-downed 293T cell. Resulting infected MT-4 cell, GFP positive, were observed by Fluorescent microscope and quantified by FACS analysis. The results are representative of four independent experiments. The means ± standard deviations of four independent experiments are shown. Statistical significance was decided by one-way analysis of variance (ANOVA); ***P < 0.001 (B) and ****P < 0.0001 (E).
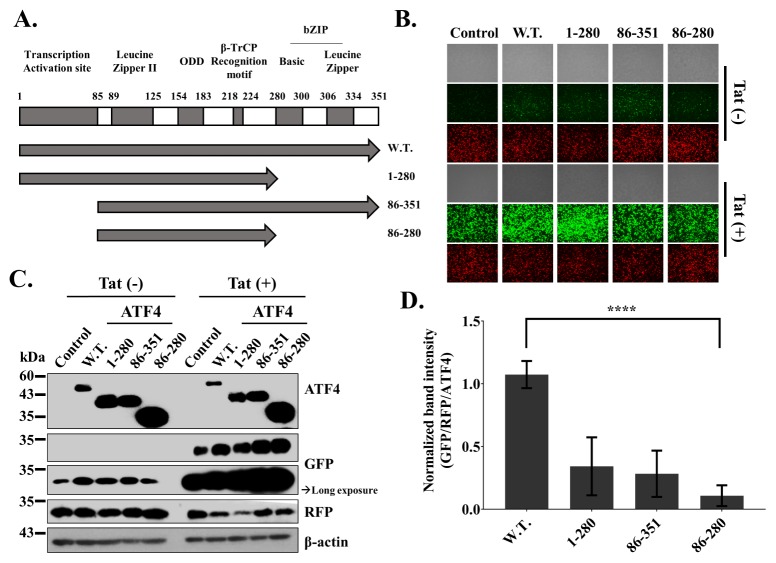
Analysis of the effect of ATF4 and its derivatives on HIV-1 LTR activation
To investigate the functional roles of ATF4 in the HIV-1 life cycle, we generated several derivative constructs of ATF4 (Fig. 2A). To determine the ATF4 domains that are functionally important in HIV-1 LTR-mediated transcription, we employed a previously described EGFP reporter gene assay (22). The expression of the EGFP reporter gene was regulated by the U3 promoter derived from HIV-1. We transiently transfected 293T cells with U3RU5/EGFP, Tat control vector, RFP expression vector as transfection control, Flag/ATF4 derivatives, or Flag control vector for 24 h. Subsequently, we analyzed the GFP and RFP signals from the transfected cells using an inverted fluorescent microscope. The levels of transcriptional activity in cells transfected with ATF4 derivatives were compared to those transfected with a Flag/Empty vector. All of the transfected cells showed nearly the same intensity of red fluorescence, indicating similar levels of transfection efficiency. Although the basal activity of the HIV-1 U3 promoter without HIV-1 Tat was very low, resulting in a weak GFP signal, the expression of wtATF4 increased relatively high GFP signal, as indicated in Fig. 2. A previous study showed that wild type ATF4 increases HIV-1 LTR activity in the presence of Tat (22). The data obtained in the present study is consistent with this result. In the presence of Tat, wtATF4 and the ATF4 derivatives 86-351 and 86-280 showed significant increases in GFP expression. However, GFP expression was much higher in cells transfected with ATF4 1-280 than in cells transfected with other ATF derivatives (Fig. 2B). To precisely compare the effects of the ATF4 derivatives on LTR activity, we conducted western blot analysis and normalized the density of the band corresponding to GFP to those of the bands corresponding to the internal control (RFP) and the ATF4 derivatives (GFP/RFP/ATF4). The result of this analysis showed that ATF4 1-280, 86-351, and 86-280 decreased LTR activity by approximately 40, 70, and 90%, respectively, relative to the LTR activity in cells with wild type ATF4 in the presence of Tat (Fig. 2C, D). By comparing the HIV-1 LTR activity of various ATF4 deletion mutants, we found that amino acids 1-85, which comprise the transcription activation site (7), and amino acids 281-351, which comprise a DNA-binding motif (6, 10), may be responsible for HIV-1 LTR activation. It is well known that p300 interacts with ATF4 and HIV-1 Tat (7, 26). Additionally, it has been shown that p300/CBP and HIV-1 Tat have a synergistic effect on the transcriptional activation of the HIV-1 promoter (27). Thus, the p300 interaction site of ATF4 would have an important functional role in HIV-1 LTR activation, possibly by interacting with p300 and Tat. Members of the ATF/CREB protein family are known to bind to the HIV-1 LTR sequence and regulate gene expression (28). The C-terminal bZIP domain of ATF4 may contribute to the ability of ATF4 to bind to and regulate the expression of the HIV-1 LTR sequence. These results suggest that both ends of ATF4 contribute to HIV-1 LTR-driven transcriptional activation.
Fig. 2.
Analysis of the effect of ATF4 and its derivatives on HIV-1 LTR activation (EGFP Reporter gene). 250 ng of U3RU5/EGFP and 100 ng of pCMV-myc/Tat (transactivator) or pCMV-Myc with 1 μg of pCMV-Flag/ATF4 derivatives or pCMV/Flag and 100 ng of RFP expression plasmid (pDs-red expression N1, internal control) were co-transfected into 293T cells for 24 hrs. (A) Cloning scheme of ATF4 derivatives. Amino acid numbers of the transcription activation/p300 interaction site, Zipper II domain, oxygen-dependent degradation domain (ODD), β-TrCP recognition motif, and basic and leucine zipper domain (bZIP) is designated at the top. Derivatives of ATF4 were exhibited at the bottom. (B) Images of 293T cells were detected by the inverted fluorescent microscope. (C) Western blot analysis. (D) Normalized relative band intensity of Fluc by RFP as an internal control and ATF4 derivatives. The results are representative of five independent experiments. The means ± standard deviations of five independent experiments are shown. Statistical significance was decided by one-way analysis of variance (ANOVA); ****P < 0.0001.
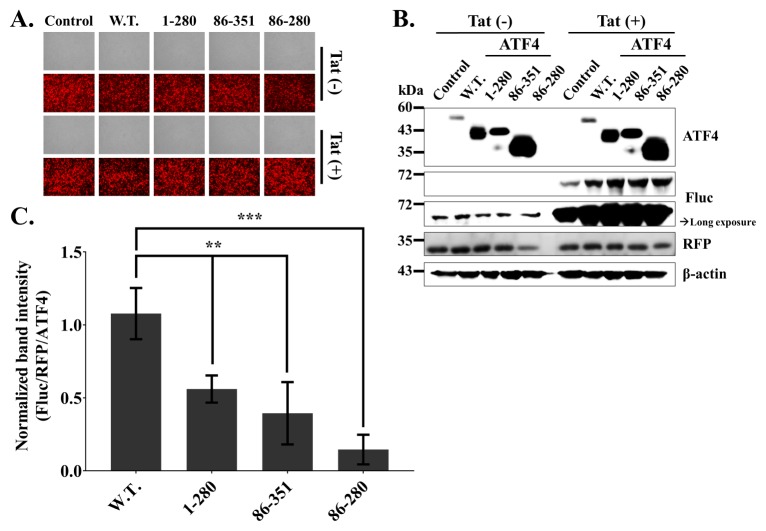
The N-terminal and C-terminal domains of ATF4 are required for HIV-1 LTR activation
To further confirm the effect of the ATF4 derivatives on HIV-1 LTR-mediated transcription activation, we used a firefly luciferase reporter gene system. As shown in Fig. 2, the expression of the reporter gene is under the control of the HIV-1 U3 promoter. We transiently transfected U3RU5/Fluc (Firefly luciferase), Tat control vector, RFP expression vector, Flag/ATF4 derivatives, or Flag-tagged control vector into 293T cells for 24 h. The levels of transcriptional activity of the cells transfected with ATF4 derivatives were compared to those of cells transfected with the Flag/Empty vector. All transfected cells showed similar intensities of red fluorescence, indicating similar levels of transfection efficiency (Fig. 3A). Western blot analysis (Fig. 3B) revealed almost the same results as shown in Fig. 2B. As shown in Fig. 2C, we normalized the intensity of the band corresponding to firefly luciferase to those of the bands corresponding to the internal control (RFP) and ATF4 derivatives (Fluc/RFP/ATF4). These data indicate that the N-terminal domain of ATF4 contributes to LTR-mediated transcriptional activation.
Fig. 3.
The N-terminal and C-terminal domains of ATF4 are required for HIV-1 LTR activation (Firefly luciferase reporter gene). 250 ng of U3RU5/Fluc (Firefly luciferase) and 100 ng of pCMV-myc/Tat (Transactivator) or pCMV-Myc with 1 μg of pCMV-Flag/ATF4 derivatives or pCMV/Flag and 100 ng of RFP expression plasmid (pDs-red expression N1, internal control) were co-transfected into 293T cells for 24 hrs. (A) Images of 293T cells were detected by the inverted fluorescent microscope. (B) Western blot analysis. (C) Normalized relative band intensity of Fluc by RFP as an internal control and ATF4 derivatives. The results are representative of three independent experiments. The means ± standard deviations of three independent experiments are shown. Statistical significance was decided by one-way analysis of variance (ANOVA); **P < 0.01 and ***P < 0.001.
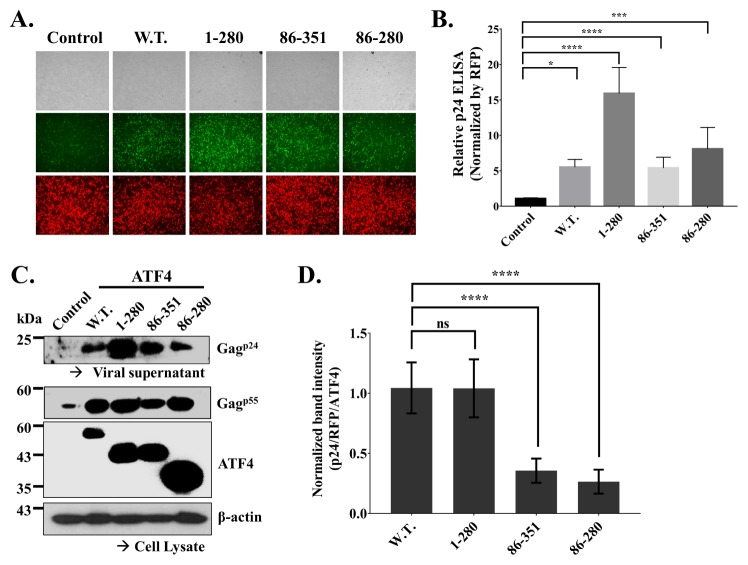
The N-terminal domain of ATF4 strongly affects the late phase of HIV-1 production
We then examined the effect of ATF4 on HIV-1 production using proviral DNA (pNL4-3/EGFP) as described previously (26). In this system, viral replication and expression can be detected by observing the EGFP signal in the cells transfected with the provirus. As shown in Fig. 3B, all transfected cells showed nearly the same intensity of red fluorescence, indicating equal transfection efficiencies. Cells transfected with ATF4 derivatives had higher GFP intensities than cells transfected with the control (Fig. 4A). We then assessed virus production by measuring the amount of HIV-1 p24 antigen in constant volumes of viral supernatants from transfected cells through an enzyme-linked immunosorbent assay (ELISA). The measured amounts of p24 were normalized against the transfection efficiency (RFP) (Fig. 4B). Cells transfected with ATF4 derivatives had much higher p24 levels than cells transfected with the control. In particular, cells transfected with ATF4 1-280 had the highest amount of p24. We then conducted western blot analysis to compare the levels of p24 in the viral supernatants and p55 in the cell lysates. The results of the western blot analysis of the viral supernatants were consistent with the results of the ELISA. As shown in Figs. 1–3, the protein expression levels of the ATF4 derivatives varied (Fig. 4C). Therefore, we needed to compare the precise effects of the ATF4 derivatives on HIV-1 production in the same conditions. To exactly compare the virus production levels of the cells transfected with different ATF4 derivatives, we normalized the protein expression levels (p24/RFP/ATF4). The normalized levels were then compared to those of cells transfected with wtATF4. In Fig. 4D, cells transfected with ATF4 86-351 and 86-280 had reduced relative p24 levels; however, cells transfected with ATF4 1-280 did not. Although a previous study showed that ATF4 without a DNA-binding function caused a decrease in MCMV growth (15), ATF4 with a deleted bZIP domain did not cause a decrease in HIV-1 production. This implies that the role of the transcription activation domain in HIV-1 production is more essential than that of the bZIP domain.
Fig. 4.
The N-terminal domain of ATF4 strongly affects the late phase of HIV-1 production. 250 ng of pNL4-3/GFP with 100 ng of RFP expression plasmid (pDs-red expression N1, internal control) and 1 μg of pCMV/Flag or pCMV-Flag/ATF4 derivatives were co-transfected into 293T Cells for 24 hrs. (A) Images of 293T cells were detected by the inverted fluorescent microscope. (B) Virus titer of viral soup was determined by p24Gag ELISA then normalized by RFP used as a control of transfection efficiency. (C) Western blot results of cell lysate and viral supernatant were shown. (D) Normalized relative band intensity of p24 by RFP as an internal control and ATF4 derivatives. The means ± standard deviation of five independent experiments is shown. Statistical significance was decided by one-way analysis of variance (ANOVA); ****P < 0.0001 (B, D), ***P < 0.001 (B), *P < 0.05 (B) and ns > 0.05 (D).
MATERIALS AND METHODS
Plasmid construction
The pCMV/Flag-ATF4 plasmids (W.T., 1-280, 86-351, 86-280) used in this study and the primers used to construct them are listed in Fig. 1 and Supplementary Table 1, respectively. The wtATF4 DNA fragments were amplified using PCR and digested with Bgl II and Eag I. Backbone DNA was digested with Bgl II. The inserts and backbone DNA were then treated with Klenow polymerase and then ligated.ATF4 mutants were constructed as follows: ATF4 1-280, 86-351 and 86-280 DNA fragments were generated via PCR amplification using pCMV/Flag-ATF4 as a template. The primers used, which are listed in Supplementary Table 1, were chemically synthesized (COSMO Genetech Co., Republic of Korea). PCR products and Flag/NC (backbone DNA) were digested with Bgl II and Hind III. The NC region in the Flag/NC backbone were removed and replaced with the PCR products. The plasmids pNL4-3/GFP (29) and U3RU5/Fluc (U3RU5 is referred to as LTR in a previous study (30)) were prepared and used for transfection as described previously. U3RU5/EGFP was constructed as follows: EGFP was amplified via PCR using U3R/EGFP (30) as a template and the primers listed in Supplementary Table 1. The PCR products and U3RU5/Fluc were then digested with NcoI and XbaI. The Fluc fragments in the U3RU5/Fluc backbone were removed and replaced with the inserts.
Cell culture, transfection, and reinfection
293T and MT4 cells were maintained in DMEM (Dulbecco’s modified Eagle’s medium) and RPMI (Roswell Park Memorial Institute Medium)-1640 respectively. Both culture media were supplemented with 10% fetal bovine serum (TCB, USA) and penicillin and streptomycin (GIBCO, Carlsbad, CA). The cells were incubated at 37°C in 5% CO2.
293T cells were transfected with plasmids using jetPEI (Polyplus-Transfection, France) following manufacturer's protocols. After a 24 h incubation, cells were harvested for further analysis. For the ATF4 knockdown, 293T cells were transfected with siATF4 or ntsiRNA (Invitrogen, USA) using lipofectamine following the manufacturer’s instructions (Invitrogen, USA). For the reinfection assay, viral supernatants were harvested, and the same volume of viral supernatants was used to re-infect MT4 cells. At 72 h post-infection, GFP expression was measured through FACS analysis.
Luciferase assay
Firefly luciferase activity was measured 24 h after transfection. The protein concentrations of the crude lysates were determined using the Bradford assay (BioRad, USA). The luciferase assay was performed using the Luciferases® Reporter Assay System (Promega, USA) and a luminometer (Infinite F200, TECAN) following the manufacturers’ protocols. Relative firefly luciferase expression levels were determined by normalizing the values against RFP expression levels. The results are presented as the mean ± standard deviation of three independent experiments (31).
Fluorescence and fluorometry
GFP- and RFP-positive 293T cells were observed using an inverted fluorescence microscope (IX71, Olympus, Japan) and quantified via fluorometry (Infinite F200, TECAN). Transfected cells were observed using Axiovert™ software connected to a fluorescent microscope (magnification: 100×). GFP and RFP intensity were measured 24 h after transfection via fluorometry (Infinite F200, TECAN) according to the manufacturer's protocols. Relative GFP expression levels were determined by normalizing the values against RFP expression levels.
Western blot analysis
Western blot was performed according to the protocols described in Molecular Cloning: A Laboratory Manual (Sambrook and Russell, 2001). Each sample was subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on a 12% acrylamide gel and transferred electrophoretically to a PVDF membrane. The primary antibodies used were anti-Flag (Sigma), anti-HIV-1 p24 (Santacruz), anti-Fluc (Cal Biochem, USA), anti-glyceraldehyde 3-phosphate dehydrogenase (GAPDH), anti-beta-actin (abcam), anti-RFP polyclonal, and anti-GFP monoclonal (Clontech/Takara Bio Co, USA). The secondary antibodies used were HRP-goat anti-mouse IgG conjugate and HRP-goat anti-rabbit IgG conjugate (Zymed, USA). Specific protein bands were developed using LAS-4000 (Fujifilm, Japan). The density of each band was analyzed using the software Image J (32).
HIV-1 p24 ELISA
The amounts of p24 in each sample were determined using an HIV-1 p24 ELISA kit following the manufacturer’s protocol (Xpressbio). Coated monoclonal antibodies against p24 epitopes were used in this kit to detect p24.
Statistical analysis
Statistical analysis was performed using one-way analysis of variance (ANOVA) with GraphPad Prism software. The p-value of <0.05 was considered statistically significant.
Supplementary Information
Acknowledgment
This work was supported by a research grant (2016R1D1A1A02937509 and 2017R1A5A1015366) from the National Research Foundation of Korean Ministry of Science and Technology, and by a grant (HI16C2095) from the Korea Health Industry Development Institute funded by the Ministry of Health and Welfare.
Footnotes
CONFLICTS OF INTEREST
The authors have no conflicting interests.
REFERENCES
- 1.Ameri K, Harris AL. Activating transcription factor 4. Int J Biochem Cell Biol. 2008;40:14–21. doi: 10.1016/j.biocel.2007.01.020. [DOI] [PubMed] [Google Scholar]
- 2.Karpinski BA, Morle GD, Huggenvik J, Uhler MD, Leiden JM. Molecular cloning of human CREB-2: an ATF/CREB transcription factor that can negatively regulate transcription from the cAMP response element. Proc Natl Acad Sci U S A. 1992;89:4820–4824. doi: 10.1073/pnas.89.11.4820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ye J, Kumanova M, Hart LS, et al. The GCN2-ATF4 pathway is critical for tumour cell survival and proliferation in response to nutrient deprivation. EMBO J. 2010;29:2082–2096. doi: 10.1038/emboj.2010.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harding HP, Novoa I, Zhang Y, et al. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol Cell. 2000;6:1099–1108. doi: 10.1016/S1097-2765(00)00108-8. [DOI] [PubMed] [Google Scholar]
- 5.Rutkowski DT, Kaufman RJ. All roads lead to ATF4. Dev Cell. 2003;4:442–444. doi: 10.1016/S1534-5807(03)00100-X. [DOI] [PubMed] [Google Scholar]
- 6.Hai T, Curran T. Cross-family dimerization of transcription factors Fos/Jun and ATF/CREB alters DNA binding specificity. Proc Natl Acad Sci U S A. 1991;88:3720–3724. doi: 10.1073/pnas.88.9.3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lassot I, Estrabaud E, Emiliani S, Benkirane M, Benarous R, Margottin-Goguet F. p300 modulates ATF4 stability and transcriptional activity independently of its acetyltransferase domain. J Biol Chem. 2005;280:41537–41545. doi: 10.1074/jbc.M505294200. [DOI] [PubMed] [Google Scholar]
- 8.Liang G, Hai T. Characterization of human activating transcription factor 4, a transcriptional activator that interacts with multiple domains of cAMP-responsive element-binding protein (CREB)-binding protein. J Biol Chem. 1997;272:24088–24095. doi: 10.1074/jbc.272.38.24088. [DOI] [PubMed] [Google Scholar]
- 9.Schoch S, Cibelli G, Magin A, Steinmuller L, Thiel G. Modular structure of cAMP response element binding protein 2 (CREB2) Neurochem Int. 2001;38:601–608. doi: 10.1016/S0197-0186(00)00127-3. [DOI] [PubMed] [Google Scholar]
- 10.Hai TW, Liu F, Coukos WJ, Green MR. Transcription factor ATF cDNA clones: an extensive family of leucine zipper proteins able to selectively form DNA-binding heterodimers. Genes Dev. 1989;3:2083–2090. doi: 10.1101/gad.3.12b.2083. [DOI] [PubMed] [Google Scholar]
- 11.Landschulz WH, Johnson PF, McKnight SL. The leucine zipper: a hypothetical structure common to a new class of DNA binding proteins. Science. 1988;240:1759–1764. doi: 10.1126/science.3289117. [DOI] [PubMed] [Google Scholar]
- 12.Cibelli G, Schoch S, Thiel G. Nuclear targeting of cAMP response element binding protein 2 (CREB2) Euro J Cell Biol. 1999;78:642–649. doi: 10.1016/S0171-9335(99)80049-1. [DOI] [PubMed] [Google Scholar]
- 13.Isler JA, Skalet AH, Alwine JC. Human cytomegalovirus infection activates and regulates the unfolded protein response. J Virol. 2005;79:6890–6899. doi: 10.1128/JVI.79.11.6890-6899.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xuan B, Qian Z, Torigoi E, Yu D. Human cytomegalovirus protein pUL38 induces ATF4 expression, inhibits persistent JNK phosphorylation, and suppresses endoplasmic reticulum stress-induced cell death. J Virol. 2009;83:3463–3474. doi: 10.1128/JVI.02307-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qian Z, Xuan B, Chapa TJ, Gualberto N, Yu D. Murine cytomegalovirus targets transcription factor ATF4 to exploit the unfolded-protein response. J Virol. 2012;86:6712–6723. doi: 10.1128/JVI.00200-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fraser JE, Wang C, Chan KW, Vasudevan SG, Jans DA. Novel dengue virus inhibitor 4-HPR activates ATF4 independent of protein kinase R-like Endoplasmic Reticulum Kinase and elevates levels of eIF2alpha phosphorylation in virus infected cells. Antiviral Res. 2016;130:1–6. doi: 10.1016/j.antiviral.2016.03.006. [DOI] [PubMed] [Google Scholar]
- 17.Liao Y, Fung TS, Huang M, Fang SG, Zhong Y, Liu DX. Upregulation of CHOP/GADD153 during coronavirus infectious bronchitis virus infection modulates apoptosis by restricting activation of the extracellular signal-regulated kinase pathway. J Virol. 2013;87:8124–8134. doi: 10.1128/JVI.00626-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Caselli E, Benedetti S, Grigolato J, Caruso A, Di Luca D. Activating transcription factor 4 (ATF4) is upregulated by human herpesvirus 8 infection, increases virus replication and promotes proangiogenic properties. Arch Virol. 2012;157:63–74. doi: 10.1007/s00705-011-1144-3. [DOI] [PubMed] [Google Scholar]
- 19.Dunlap KM, Bartee MY, Bartee E. Myxoma virus attenuates expression of activating transcription factor 4 (ATF4) which has implications for the treatment of proteasome inhibitor-resistant multiple myeloma. Oncolytic Virother. 2015;4:1–11. doi: 10.2147/OV.S72372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fros JJ, Major LD, Scholte FE, et al. Chikungunya virus non-structural protein 2-mediated host shut-off disables the unfolded protein response. J Gen Virol. 2015;96:580–589. doi: 10.1099/vir.0.071845-0. [DOI] [PubMed] [Google Scholar]
- 21.Burnett HF, Audas TE, Liang G, Lu RR. Herpes simplex virus-1 disarms the unfolded protein response in the early stages of infection. Cell Stress Chaperones. 2012;17:473–483. doi: 10.1007/s12192-012-0324-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Caselli E, Benedetti S, Gentili V, Grigolato J, Di Luca D. Short communication: activating transcription factor 4 (ATF4) promotes HIV type 1 activation. AIDS Res Hum Retroviruses. 2012;28:907–912. doi: 10.1089/aid.2011.0252. [DOI] [PubMed] [Google Scholar]
- 23.Krebs FC, Goodenow MM, Wigdahl B. Neuroglial ATF/CREB factors interact with the human immunodeficiency virus type 1 long terminal repeat. J Neurovirol. 1997;3(Suppl 1):S28–32. [PubMed] [Google Scholar]
- 24.Krebs FC, Mehrens D, Pomeroy S, Goodenow MM, Wigdahl B. Human immunodeficiency virus type 1 long terminal repeat quasispecies differ in basal transcription and nuclear factor recruitment in human glial cells and lymphocytes. J Biomed Sci. 1998;5:31–44. doi: 10.1007/BF02253354. [DOI] [PubMed] [Google Scholar]
- 25.Gachon F, Peleraux A, Thebault S, et al. CREB-2, a cellular CRE-dependent transcription repressor, functions in association with Tax as an activator of the human T-cell leukemia virus type 1 promoter. J Virol. 1998;72:8332–8337. doi: 10.1128/jvi.72.10.8332-8337.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hottiger MO, Nabel GJ. Interaction of human immunodeficiency virus type 1 Tat with the transcriptional coactivators p300 and CREB binding protein. J Virol. 1998;72:8252–8256. doi: 10.1128/jvi.72.10.8252-8256.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ott M, Schnolzer M, Garnica J, et al. Acetylation of the HIV-1 Tat protein by p300 is important for its transcriptional activity. Curr Biol. 1999;9:1489–1492. doi: 10.1016/S0960-9822(00)80120-7. [DOI] [PubMed] [Google Scholar]
- 28.Gaynor R. Cellular transcription factors involved in the regulation of HIV-1 gene expression. Aids. 1992;6:347–363. doi: 10.1097/00002030-199204000-00001. [DOI] [PubMed] [Google Scholar]
- 29.Kim MJ, Kim SH, Park JA, et al. Identification and characterization of a new type of inhibitor against the human immunodeficiency virus type-1 nucleocapsid protein. Retrovirology. 2015;12:90. doi: 10.1186/s12977-015-0218-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jeong HW, Kim SH, Sim SY, Yu KL, You JC. The HIV-1 nucleocapsid protein does not function as a transcriptional activator on its own cognate promoter. Virus Res. 2012;163:469–475. doi: 10.1016/j.virusres.2011.11.009. [DOI] [PubMed] [Google Scholar]
- 31.Jang E, Lee HR, Lee GH, et al. Bach2 represses the AP-1-driven induction of interleukin-2 gene transcription in CD4 T cells. BMB Rep. 2017;50:472–477. doi: 10.5483/BMBRep.2017.50.9.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shin CH, Ryu S, Kim HH. hnRNPK-regulated PTOV1-AS1 modulates heme oxygenase-1 expression via miR-1207-5p. BMB Rep. 2017;50:220–225. doi: 10.5483/BMBRep.2017.50.4.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.