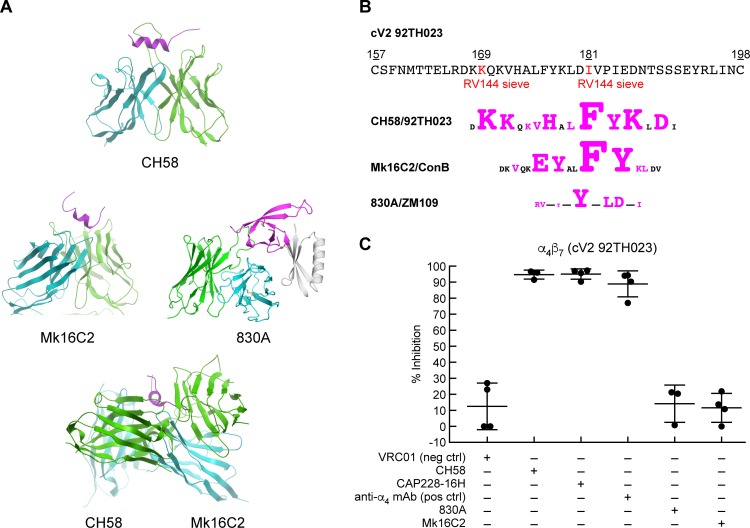
Fig 3. The structures of V2 mAbs in complex with peptides, and V2 mAb inhibition of α4β7 adhesion.
A) Crystal structure of three V2 domain mAbs, CH58 (PDB ID: 4HPO), Mk16C2 (PDB ID: 6CEZ), and 830A (PDB ID: 4YWG) in complex with V2 peptides, and a superimposition of CH58 and Mk16C2 binding to opposite sides of the same helical region of a V2 peptide. Only the Fv regions are shown with the heavy chain, light chain, and the V2 epitope colored cyan, green, and magenta, respectively. B) Amino acid sequence of the V2 domain of 92TH023. Sieve residues identified in the RV144 vaccine study are highlighted in red, along with schematics of the epitopes of CH58, Mk16C2 and 830A aligned below (with the corresponding HIV isolate listed). Residues in contact with each mAb are highlighted in magenta and the size of each amino acid is proportional to the contact surface area. C) Adhesion of RPMI8866 cells to cV2 92TH023 in the absence or presence of V2 mAbs: CH58, CAP228-16H, 830A, and Mk16C2. The mAb VRC01 is included as a nonspecific mAb control, and the anti-α4 mAb 2B4 is employed as a positive control. Adhesion was determined at OD590nm and listed as % inhibition in three or more independent experiments relative to cV2 92TH023 in the absence of any inhibitor (y-axis). Error bars indicate SD.