Figure 2. IAV-induced read-through and structural changes are NS1-dependent.
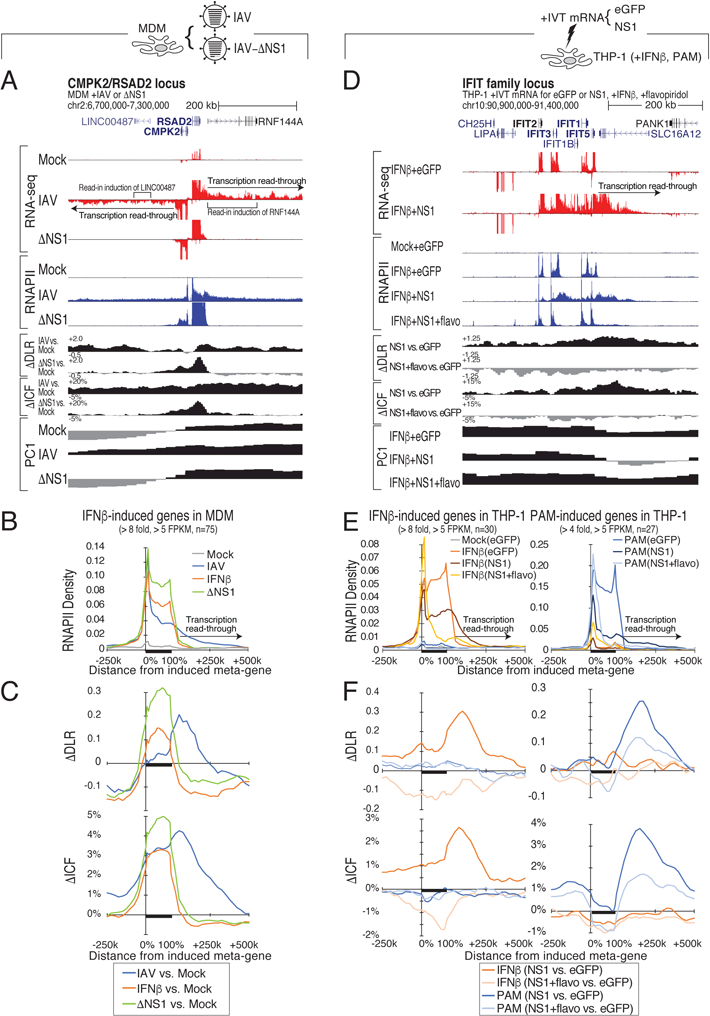
(A) Genome browser tracks of RNA-seq and RNAPII ChIP-seq read densities and chromatin compaction changes at the CMPK2/RSAD2 locus in mock-, wild-type IAV-, and IAV with NS1 truncation ΔNS1)-infected MDM. (B,C) Meta-gene plot of RNAPII ChIP-seq reads (B) and changes in chromatin compaction ΔDLR and ΔICF) (C) after IAV, ΔNS1, or IFNβ treatment of MDM at highly IFNβ-responsive genes defined in Figure 1F. (D) Genome browser tracks of RNA-seq and RNAPII ChIP-seq reads and chromatin compaction changes at the IFIT family locus in THP-1 cells expressing either EGFP or influenza NS1 protein with or without IFNβ and flavopiridol. (E,F) Meta-gene plot of RNAPII ChIP-seq reads (E) or changes in chromatin compaction ΔDLR and ΔICF) (F) in THP-1 cells expressing EGFP or NS1, treated with either IFNβ or PAM (TLR2 agonist) and flavopiridol at genes highly responsive to IFNβ or PAM stimulation in THP-1 (IFNβ: FC > 8, RNAPII FPKM > 5; 30 response genes; PAM: FC > 4, RNAPII FPKM > 5; 27 response genes, see Table S2). See also Figure S2.
