Figure 4. Intragenic chromatin loops weaken with normal transcriptional responses to cellular signaling.
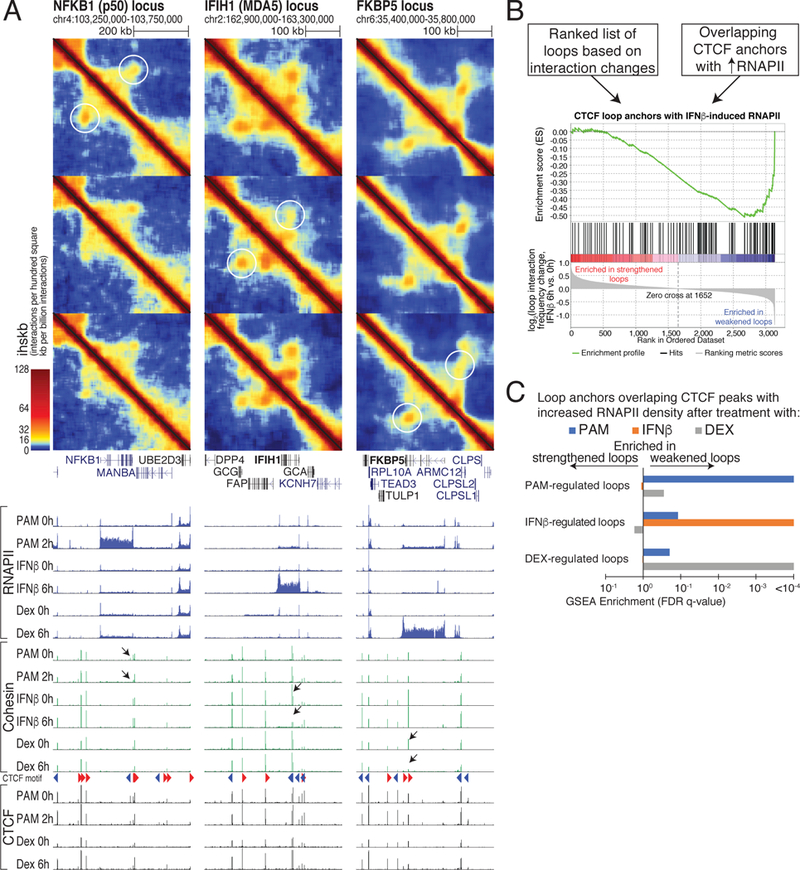
(A) Hi-C contact maps and RNAPII, cohesin, and CTCF ChIP-seq signal at loci that selectively respond to PAM (NFKB1), IFNβ (IFIH1), and dexamethasone (FKBP5) in THP-1 cells. Chromatin loops are weakened only if the corresponding gene is induced. Black arrows mark intragenic CTCF sites where cohesin is lost following transcription induction. (B) Gene Set Enrichment Analysis (GSEA) of ranked lists of chromatin loops for loop anchor overlap at CTCF sites with increased RNAPII following treatment (>1.5 fold). Loops are ranked by their log2 ratio of Hi-C interactions in treated versus control conditions. (C) Association between transcribed CTCF sites and chromatin loop changes is signaling-specific. GSEA enrichment q-values comparing each ranked list of chromatin loop changes with each set of CTCF sites with increased RNAPII density for PAM, IFNβ, and dexamethasone (Dex) treatments. See also Figures S4 and S5.
