Figure 6. Kinetic analysis of RNAPII elongation and cohesin displacement.
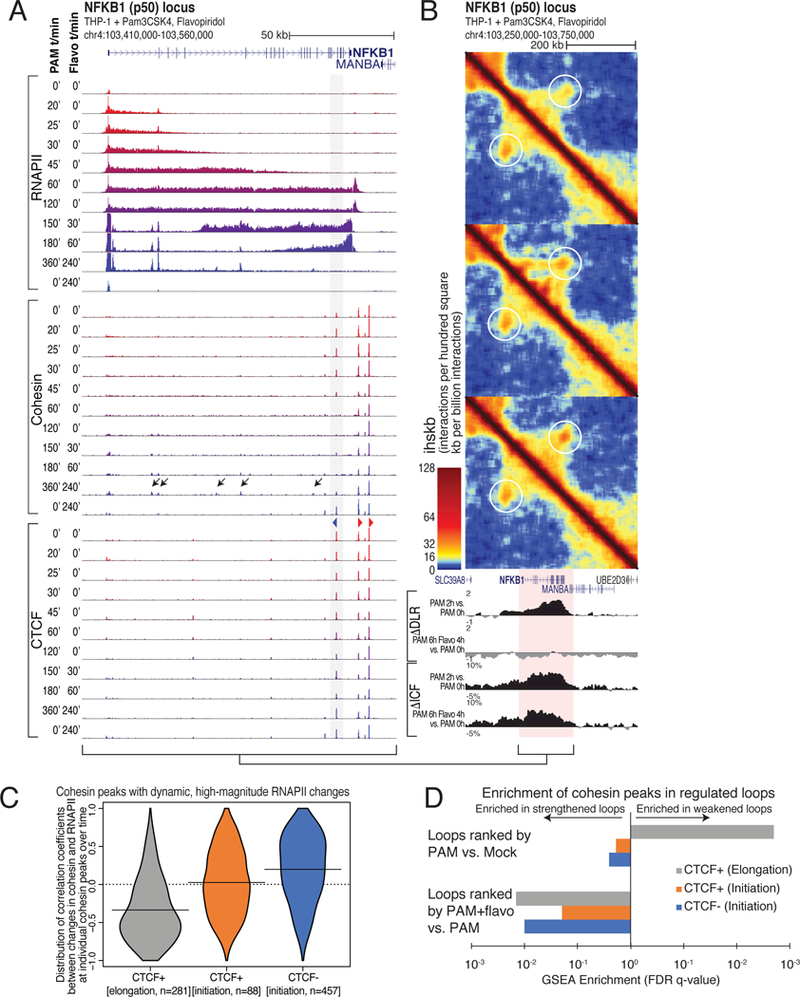
(A) RNAPII, cohesin, and CTCF ChIP-seq at the NFKB1 (p50) locus. THP-1 cells treated with PAM followed by flavopiridol for the indicated times. Flavopiridol treatment for 240 minutes is included as a control. Grey box, CTCF site that loses cohesin when transcribed by RNAPII. Black arrows mark putative PAM-specific enhancers that accumulate RNAPII and cohesin following PAM and flavopiridol treatments. (B) Hi-C contact maps of the NFKB1 locus showing the weakening of the loop formed by the CTCF site highlighted by grey box in (A). Altered interaction patterns are observed near the NFKB1 gene body in PAM+flavopiridol-treated samples. (C) Distribution of correlation coefficients between the temporal levels of RNAPII and RAD21 for each RAD21 peak exhibiting high-magnitude changes in RNAPII density (FC > 2, RNAPII FPKM > 10). Peaks were segregated based on CTCF (>10 FPKM) and whether or not they are found in ‘initiating’ or ‘elongation’ regions based on the increase or decrease in RNAPII density following flavopiridol treatment. (D) GSEA enrichment q-values for changes in loop strength overlapping with the subsets of RAD21 peaks defined in (C). See also Figure S6.
