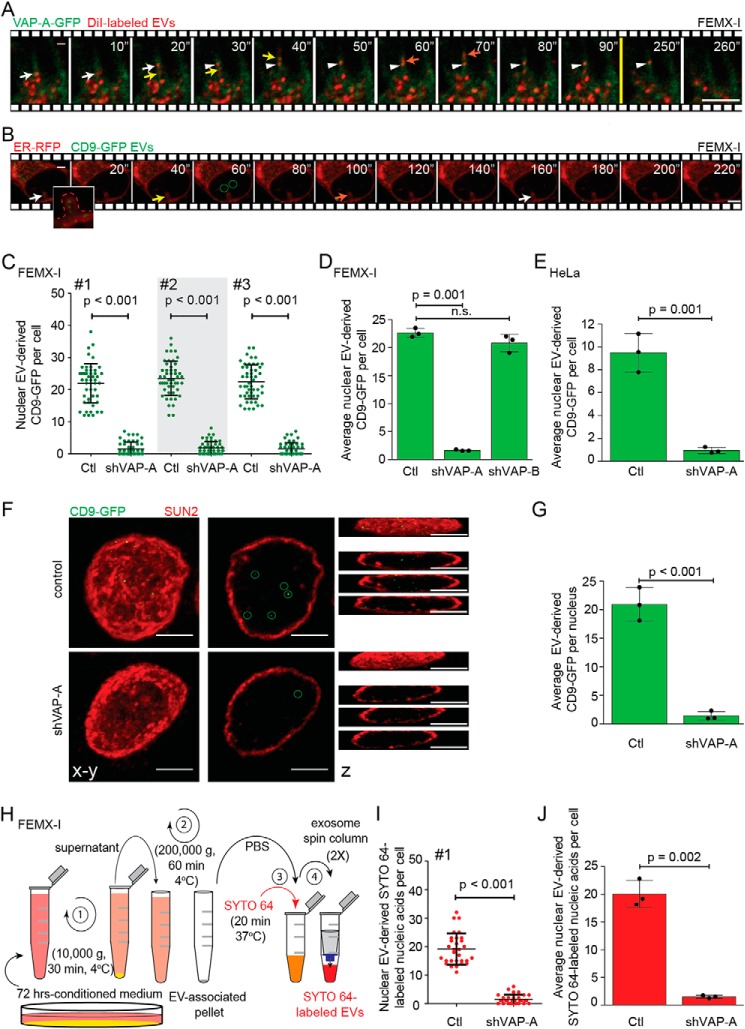
Figure 7.
VOR complex is required for nuclear transfer of EV-derived proteins and nucleic acids. A and B, FEMX-I cells expressing either VAP-A–GFP (A) or ER–RFP (B) were incubated for 5 h with DiI-labeled EVs or CD9–GFP EVs, respectively, prior to analysis by time-lapse video microscopy. Elapsed time is indicated in the top right corner. Arrows indicate the entry of late endosomes containing DiI-labeled membranes (A) or CD9–GFP (B) in NEI and the arrowhead their tether to VAP-A–GFP+ ONM (A). GFP+ signals appearing in the nucleoplasm were highlighted (B, circles). Inset in B shows enlargement of NEI (dashed line) containing discrete punctate CD9–GFP signals. Still images (A) are from Video S5. C–E, scrambled shRNA (control, Ctl) or shVAP-A/B–transfected FEMX-I (C and D) or HeLa (E) cells were incubated with fluorescent EVs derived from CD9–GFP-expressing FEMX-I cells and then double-immunolabeled for VAP-A or VAP-B and SUN2 prior to CLSM. The amount of EV-derived CD9–GFP in the nuclear compartment was quantified using Fiji software. Micrographs are presented in Fig. S8. Independent values for each cell from three independent experiments (C, #1–3) and their average from three independent experiments (D and E) are presented. F and G, nuclei of FEMX-I cells incubated with CD9–GFP+ EVs were isolated before SUN2 immunolabeling and CLSM. Composite, single x-y section and z-projections are shown (F). GFP+ signals in the nucleoplasm were highlighted (circles) and quantified (G). H, scheme of isolation of EVs from FEMX-I cells and subsequent nucleic acid staining with SYTO 64 dye. I and J, scrambled shRNA or shVAP-A–transfected FEMX-I cells were incubated with SYTO 64-labeled EVs and immunolabeled as above. Nuclear SYTO 64 signals in a given cell (I, experiment #1) and their average from three independent experiments (J) were quantified. In all cases, means ± S.D. are shown (n = 3). More than 50 (C and D) and 30 (E, I, and J) cells or 30 isolated nuclei (G) were evaluated per experiment. p values are indicated. n.s., not significant. Scale bars, 5 μm.