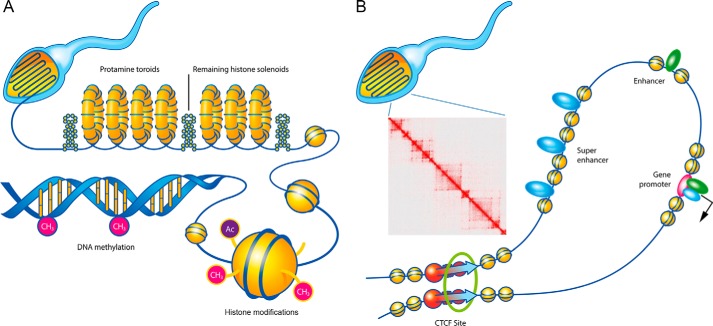
Figure 1.
Old and new views of chromatin structure in mature sperm. A, most sperm DNA was thought to be packaged in protamine toroids (yellow rings) and remaining histone octameres (yellow spheres). DNA is methylated at 5mC (pink), whereas histones are methylated or acetylated in their N-tails. B, recent results suggest the retention of more nucleosomes than previously thought in mouse sperm. These nucleosomes contain most or all histone modifications found in somatic cells (data not shown). Well-positioned nucleosomes flank TSSs and transcription factor-binding sites, including CTCF. Many enhancers and super-enhancers active in embryonic or adult tissues are already specified in the sperm epigenome. CTCF and cohesin mediate long-range chromatin interactions and establish the three-dimensional architecture of the chromatin. This is represented by a Hi-C heatmap displaying sperm chromatin organization into compartments and domains similar to those found in other cell types.