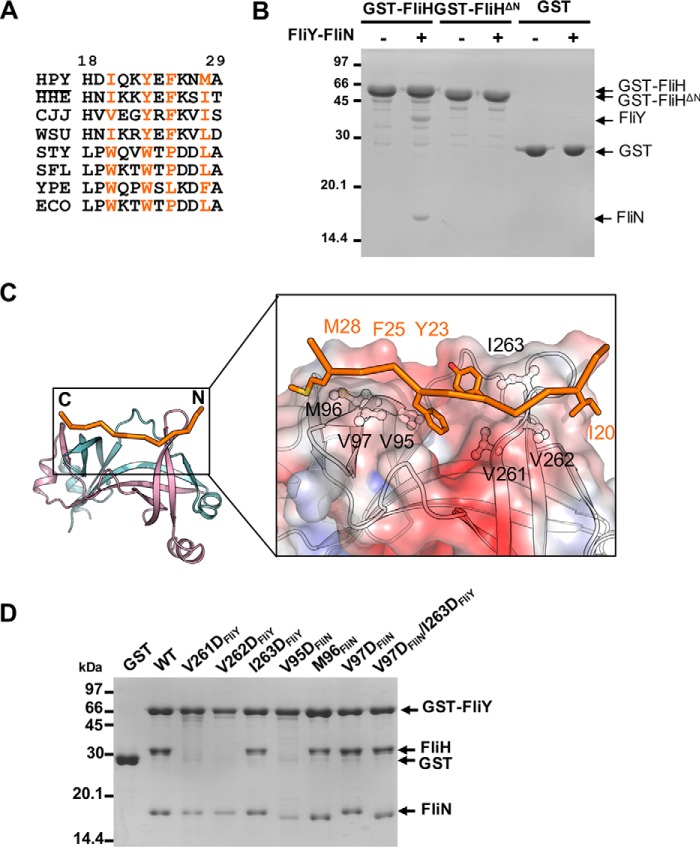
Figure 7.
Mapping the binding interface between FliH and FliY-FliN. A, excerpted T-COFFEE alignment of H. pylori (HPY) FliH with its homologues from Helicobacter hepaticus (HHE), C. jejuni (CJJ), Wolinella succinogenes (WSU), S. enterica Typhimurium (STY), Shigella flexneri (SFL), Yersinia pestis (YPE), and E. coli (ECO). The conserved hydrophobic residues are colored orange (H. pylori numbering). B, effect of FliH truncation on the binding to FliY-FliN. GST, GST-FliH, or GST-FliHΔ27 was immobilized, and the beads were incubated with purified FliY-FliN. C, model of H. pylori FliHN (residues 18–29) docked onto FliYC–FliNC. The initial model of FliHN was generated by Modeler using FliHN of S. enterica Typhimurium (PDB code 4YXC) as a template (44). The peptide was docked onto the FliYC–FliNC complex using the flexible docking protocol Rosetta FlexPepDock (55). The model with the highest score was chosen. The binding site for FliHN is shown as electrostatic surface in the boxed-in region. The molecular surface was calculated by APBS (Adaptive Poisson-Boltzmann Solver) and contoured at keT = ±3. The conserved hydrophobic residues of FliHN and FliYC–FliNC that participated in the interaction are represented as stick and ball-and-stick models, respectively. D, effect of FliY–FliN mutations on FliH interaction. The GST-FliY–FliN mutants were immobilized on resin followed by incubation with purified FliH.