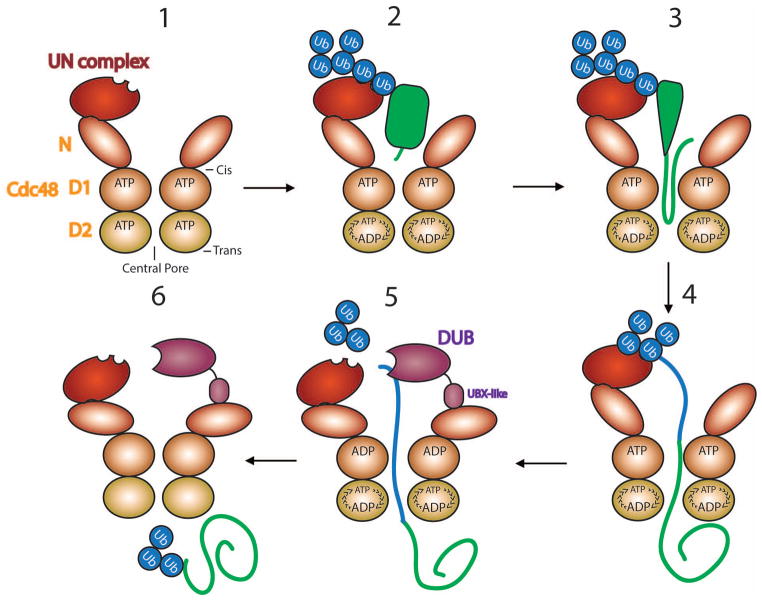
Figure 3. Substrate processing by the Cdc48 ATPase.
The scheme shows different stages. Stage 1: The Cdc48 ATPase, containing an N domain and two ATPase domains (D1 and D2), forms a hexameric, double-ring structure that associates with one copy of the Ufd1/Npl4 (UN) cofactor. The central pore and the cis- and trans-sides are indicated. Stage 2: The ubiquitin (Ub) chain attached to a substrate (in green) binds to UN. The D1 ATPases are locked in the ATP-bound state with the N domains in the up-conformation, while the activity of the D2 ATPases is stimulated. Stage 3: The substrate is pulled through the central pore, causing polypeptide unfolding. Stage 4: The substrate is moved entirely to the trans-side of the ATPase ring. Ubiquitin is also unfolded and follows the substrate (blue line). Stage 5: ATP hydrolysis in D1 causes movement of the N domains into the down-conformation, allowing a DUB to trim the ubiquitin chain. The DUB Otu1 binds through its UBX-like domain to the N domain. Stage 6: Ubiquitin molecules emerging at the trans-side presumably refold, allowing the substrate to be recognized by shuttling factors and the proteasome.