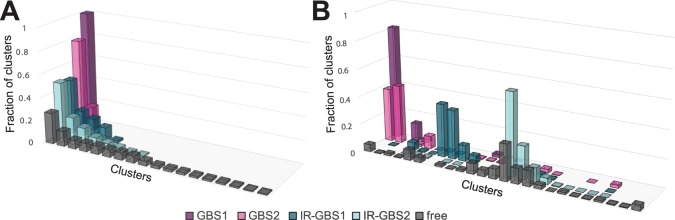
Figure 7.
Cluster analysis for lever arm conformations. (A) Clustering of individual MD trajectories shows trends in the conformational space sampled in different states of GR. For each molecule clusters are ordered by decreasing frequency. (B) Combined clustering of the duplicate trajectories for DNA-free GR DBD (molecule C), 4HN5, and 3G99 shows which clusters are shared between the different molecules. The results show that the two monomers bound to a GBS spend ~40% of their time in the same conformation. Further significant overlap is observed between free GR and the low affinity binding site within the IR-GBS complex (IR-GBS2). Molecule IR-GBS1 does not have significant overlap with any other molecules.