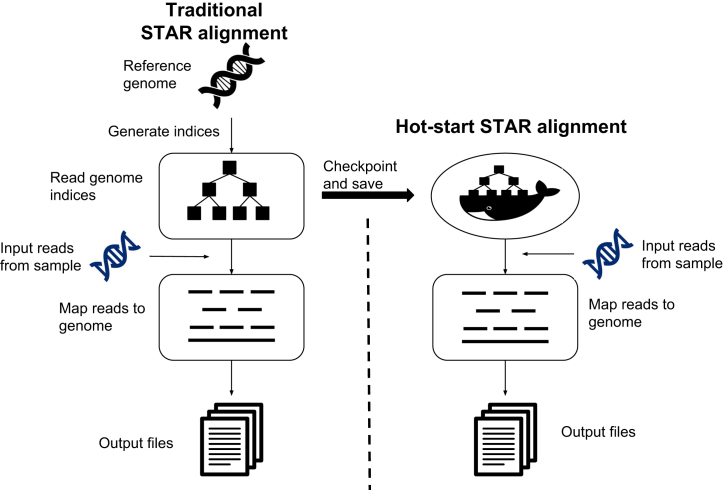
Figure 1:
An overview of our approach with and without checkpoints. The left panel shows the two steps of the STAR aligner [14, 15] after the generation of indices. The right panel shows our approach using the CRIU tool that freezes a running container and saves the checkpoint as a collection of files on disk after the genome indices are generated using the reference genome. Our “hot-start” containers use these saved files to restore the application and map the reads from the experimental sample data to the reference.