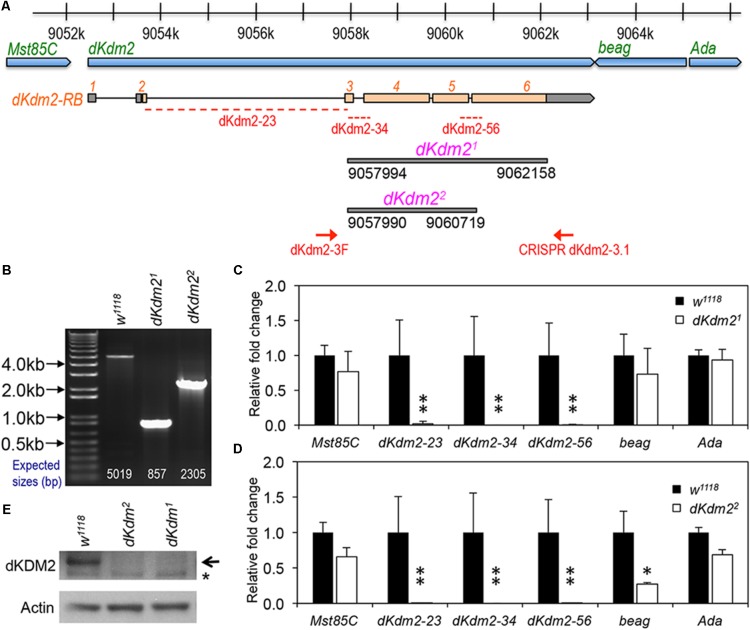
FIGURE 1.
Generation and characterization of the dKdm2 null alleles. (A) Schematic drawing of the dKdm2 locus and its neighboring genes, note the regions that are deleted in dKdm21 and dKdm22 and the corresponding breakpoints. (B) Validation of the dKdm2 alleles using PCR and genomic DNA. The positions of the primers (“dKdm2-3F” and “CRISPR dKdm2-3.1”) are shown in (A). (C,D) The mRNA levels of dKdm2 and the neighboring genes dKdm2, Mst85C, beag, and Ada in the third instar larvae are measured by qRT-PCR assay. The positions of the primers for three regions within the dKdm2 locus are shown in (A). ∗P < 0.05, ∗∗P < 0.01 based on Student’s t-tests (one-tailed). (E) The protein levels of dKDM2 in the third instar (L3) wandering larvae were analyzed by Western blot. The non-specific bands are marked with ‘∗,’ and anti-actin was used as the control.