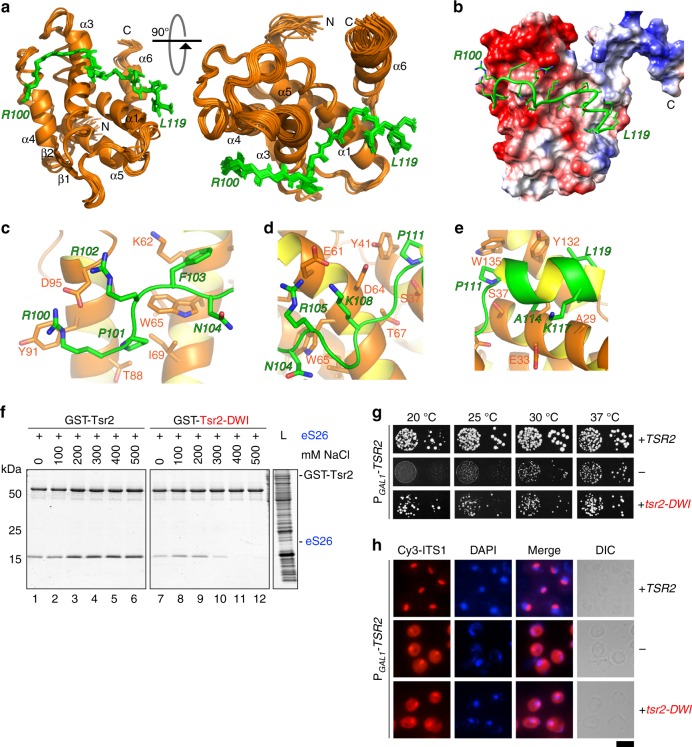
Fig. 3.
Molecular interactions between Tsr2 and ESS2. a Superposition of the 20 lowest-energy conformers representing the solution structure of Tsr2-N in complex with ESS2. b Coulomb potential projected onto the solvent accessible surface of Tsr2 with the backbone of ESS2 represented by tube; sidechains are presented by sticks with oxygens red, carbons green, nitrogens blue, and hydrogens white. The potential due to Tsr2 is represented by red for negative values (acidic residues) and blue for positive values (basic residues). The C-terminus of Tsr2-N is visible as the blue arm extending to the right. c–e Closeup of various interactions between residues of TSR2-N (orange letters label sidechains shown in stick representation) and ESS2 (green italic letters). Sidechain atoms are colored as in b. f Tsr2DWI mutant shows impaired interaction with eS26 in vitro. GST-Tsr2 or GST-Tsr2DWI was incubated with recombinant eS26 at the indicated salt concentrations before pull-down. L = input (1:10 diluted). Results from in vitro binding were quantified using ImageJ. g Tsr2DWI mutant impairs yeast growth. The conditional PGAL1-TSR2 strain was transformed with empty vector or vector with TSR2 WT or tsr2DWI. Transformants were spotted in 10-fold dilutions on repressive glucose containing media and grown at indicated temperatures for 2–4 days. h Tsr2DWI mutant cells accumulate immature 20S pre-rRNA in the cytoplasm. PGAL1-TSR2 cells transformed with WT TSR2 or tsr2DWI were grown at 30 °C in glucose containing media to the mid-log phase. Localization of 20S pre-rRNA was analyzed by FISH. Scale bar = 5 µm