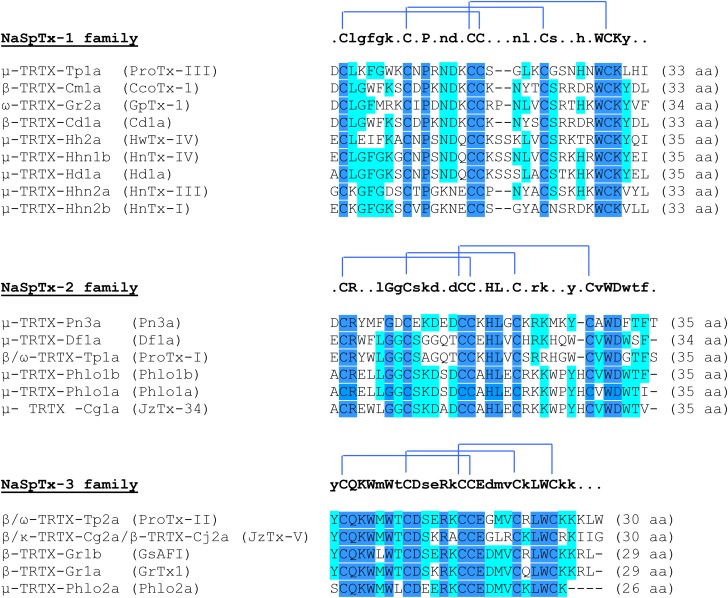
FIGURE 5.
Sequence alignment obtained by Multalin (version 5.4.1) of different potential analgesic toxins sorted by NaV spider toxins families, using their UniProtKB identifiers. The consensus sequence is shown above each alignment, with the disulfide bond connectivity. In dark blue, highly conserved amino acid residues (100%), and in light blue, poorly conserved amino acid residues (>50%). The Greek letter(s) before the toxin name is associated to its type of action: μ for NaV channel inhibition, β for shift in the voltage-dependence of NaV channel activation, ω for CaV channel inhibition, and κ for KV channel inhibition. ProTx, protoxin; HnTx, hainantoxin; CcoTx, ceratotoxin; HwTx, huwentoxin; JzTx, jingzhaotoxin; aa, amino acid residues; NaSpTx, spider NaV channel toxin.