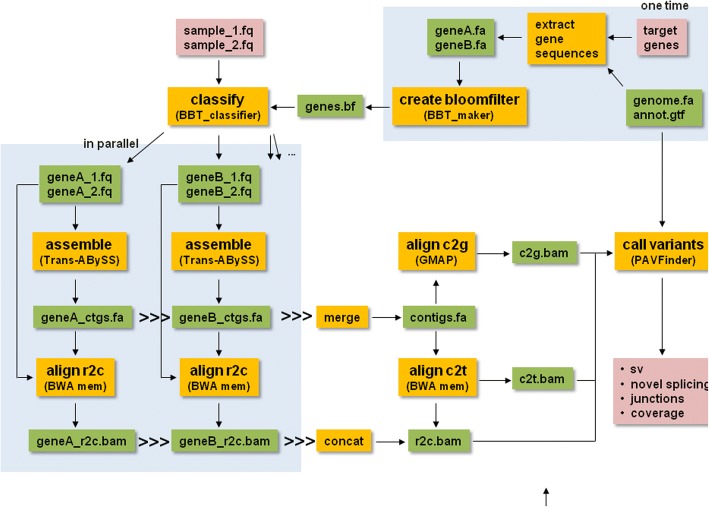
Fig. 1.
TAP Pipeline. A Bloom filter is generated from reference transcript sequences of a target list and then applied on full transcritpome RNA-seq sequences to extract gene-specific read pairs. Reads classified to each target are segregated into separate bins and assembled using two k-mer values independently in parallel. Contigs from each k-mer assembly of each gene are merged and extracted reads are aligned to them (r2c). Gene-level assemblies are combined into a single file and aligned to the genome (c2g) and transcriptome (c2t). PAVFinder uses the c2g and c2t alignments together with contig sequences and annotation (reference sequences and gene models) to identify structural variant and novel splicing events. r2c alignments are used for determining event support and coverage estimation