Fig 1.
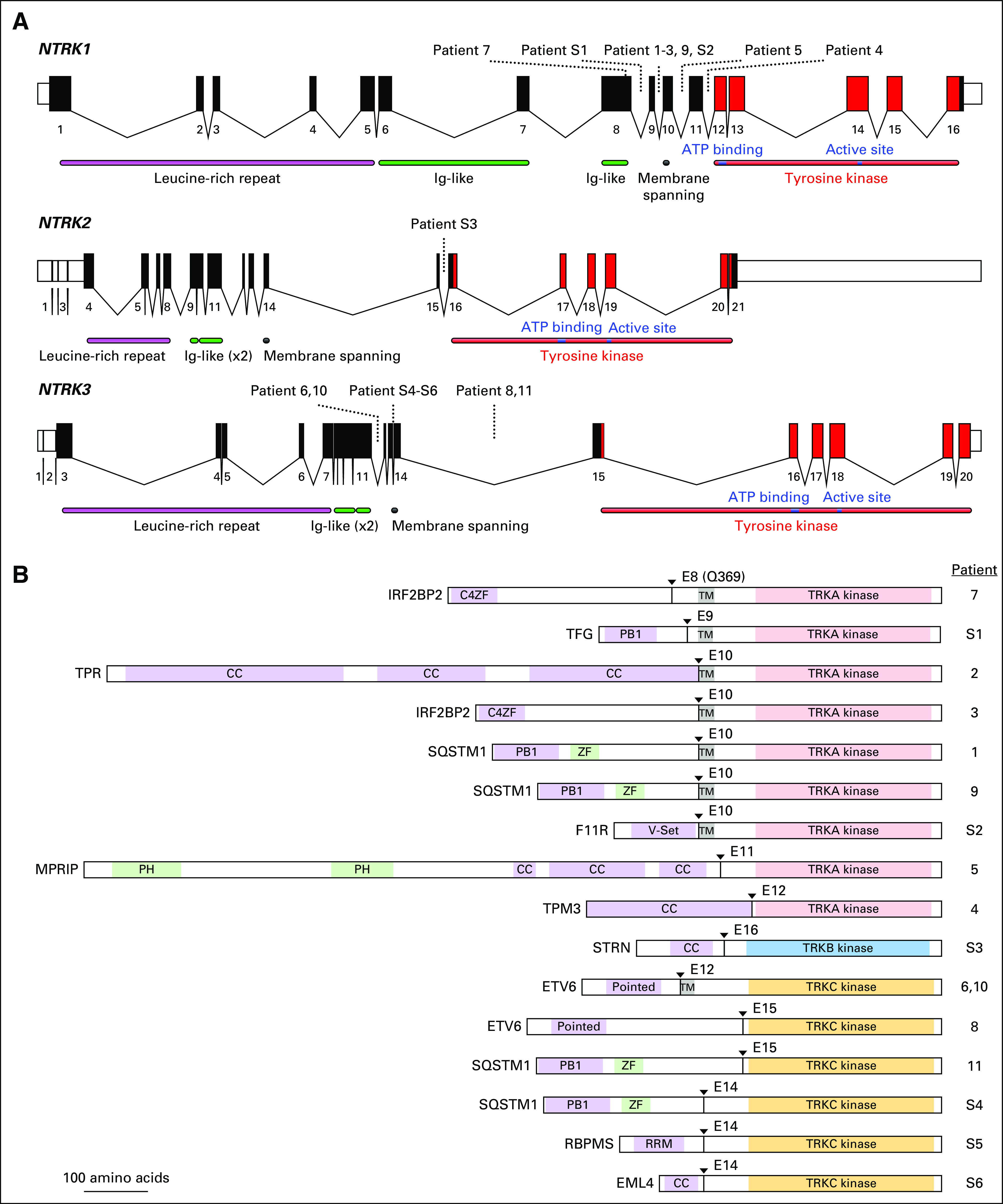
(A) Schematic of the human NTRK loci. Exon numbers are shown below their respective boxes for reference sequence NTRK1 transcript variant 1 (NM_001012331.1), NTRK2 transcript variant a (NM_006180.4), and NTRK3 transcript variant 1 (NM_001012338.2). Fusion breakpoints are shown as dotted lines for the indicated patients. Patient 7 has an exonic breakpoint; all other breakpoints are intronic. Note that exons are drawn at a larger scale than introns and that introns are not drawn to the same scale for each gene (NTRK1 locus is approximately 21 kilobases [kb], NTRK2 is approximately 358 kb, and NTRK3 is approximately 384 kb). (B) Schematic of predicted fusion protein products (see also Tables 2 and 3). Triangles and E notation indicate the fusion breakpoints and subsequent TRK exon. Purple-shaded domains are those predicted or shown to induce dimerization in the fusion partner (C4ZF, C4 zinc finger; PB1, Phox and Bem1p interaction domain; CC, coiled coil; Pointed, sterile alpha motif [SAM]/helix loop helix [HLH] oligomerization domain; RRM, RNA recognition motif). Green-shading indicates domains are other annotated sequence features (ZF, zinc finger; PH, pleckstrin homology). Gray shading indicates the transmembrane domain (TM). The kinase domains of TRKA/B/C are indicated and shown in light red, blue, and gold, respectively. Proteins are drawn to scale (MPRIP-NTRK1 fusion = 1,332 amino acids). Ig, immunoglobulin.
