Figure 3: Spatial analysis reveals a hierarchy of organization of tumor and immune cells.
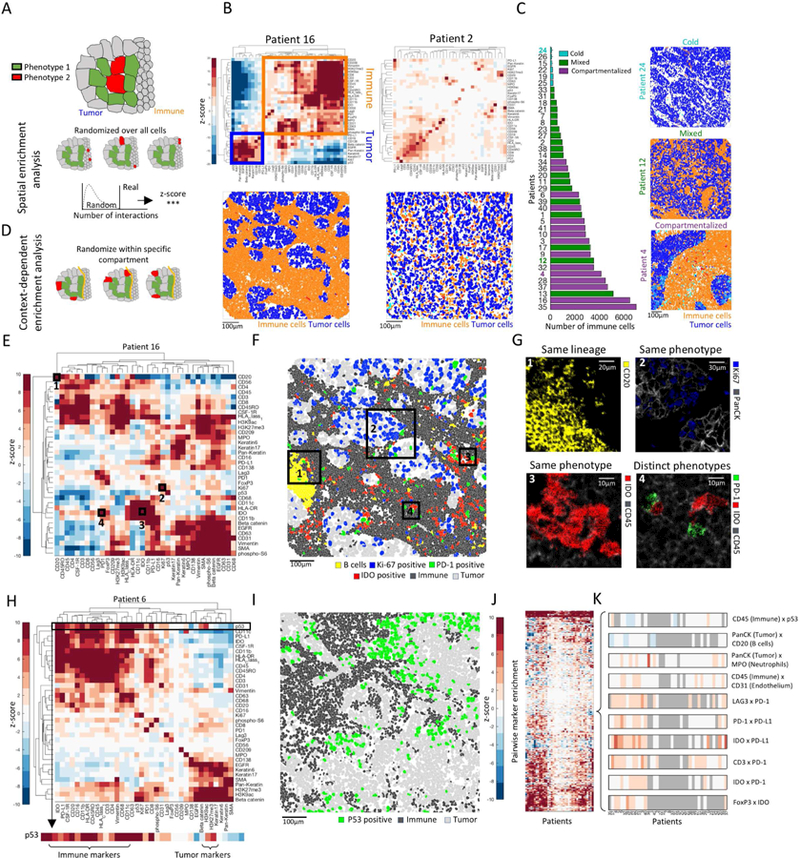
(A) Assessment of enriched proximity/distance between two cell types, red and green. Location of red cells is randomized to generate a null distribution of red-green interactions. The actual number is compared to the distribution to calculate a z-score. (B) Top: Heat maps depicting spatial enrichment z-scores between pairs of proteins. Orange and blue boxes mark immune-and tumor-related proteins, respectively. Bottom: Pseudo coloring of immune and tumor cells. (C) Left: Patients, sorted by the number of immune cells and colored by their spatial architecture. Right: Pseudo coloring of immune and tumor cells in three patients. (D) Context-dependent spatial enrichment (CDSE) analysis controls for hierarchical tissue organization. The location of red cells is randomized only within the tumor compartment. (E) Heatmap of CDSE z-scores between pairs of proteins in patient 16. Squares are highlighted in F and G. (F) Pseudo-coloring of cell populations highlighted in E and G. (G) Protein staining in boxed regions in F. (H) Heatmap of CDSE z-scores between pairs of proteins in patient 6. Zoom-in on p53 shows its proximity to immune cells. (I) Pseudo-coloring of patient 6 showing increased proximity of p53+ tumor cells to immune cells. (J) CDSE z-scores, grouped for all patients. Patients are ordered as in K. (K) Zoom-in on a few interactions in J displaying variability (CD45xp53), depletion (Pan-KeratinxCD20) or enrichment (rest) across patients. Gray denotes patients with less than 10 positive cells for either maker.
