Fig. 1.
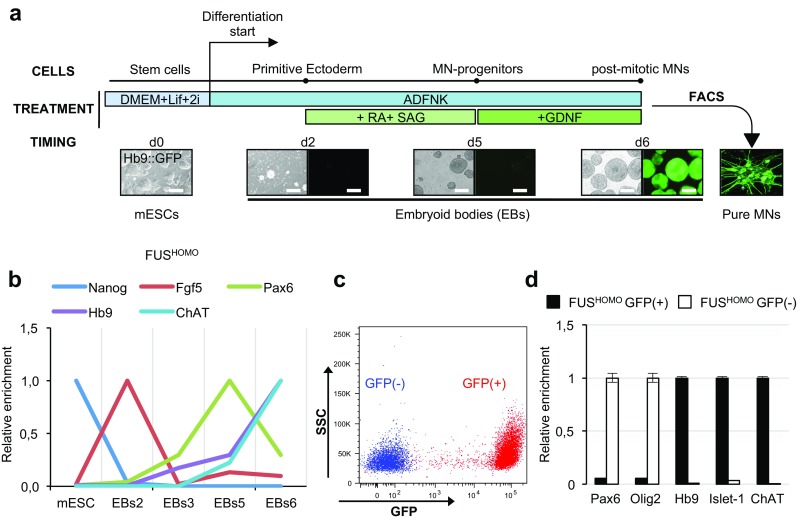
In vitro differentiation of mESC into spinal MNs. a. Schematic overview of the differentiation protocol: cell types, experimental treatments, and timing are indicated (d = day, RA = retinoic acid, SAG = smoothened agonist, FACS = fluorescence-activated cell sorting, GDNF = glial cell-derived neurotrophic factor, ESCs = embryonic stem cells, EBs = embryoid bodies, MNs = motor neurons). See text for details. Scale bar: 200 μm. b qRT-PCR profiling of stemness, primitive ectoderm, and neural/MN markers along differentiation of FUSHOMO mESCs to MNs. Cell types/differentiation days (2 to 6) are indicated on the x-axis. For each marker analyzed (indicated above), the expression peak is set as 1. Results are expressed in arbitrary units, relative to Atp5o as internal standard. c mESCs differentiated to MNs were isolated by FACS based on GFP expression level. Upon sorting, FUSHOMO GFP(−) (blue) and GFP(+) (red) cells were checked for purity. The figure shows the overlay of the purified populations. d qRT-PCR analysis of neural/MN progenitors and MN markers in sorted FUSHOMO GFP(+) (black bars) and GFP(−) cell populations (white bars). For each marker analyzed (indicated below), the expression peak is set as 1. Results are expressed in arbitrary units, relative to Atp5o as internal standard
