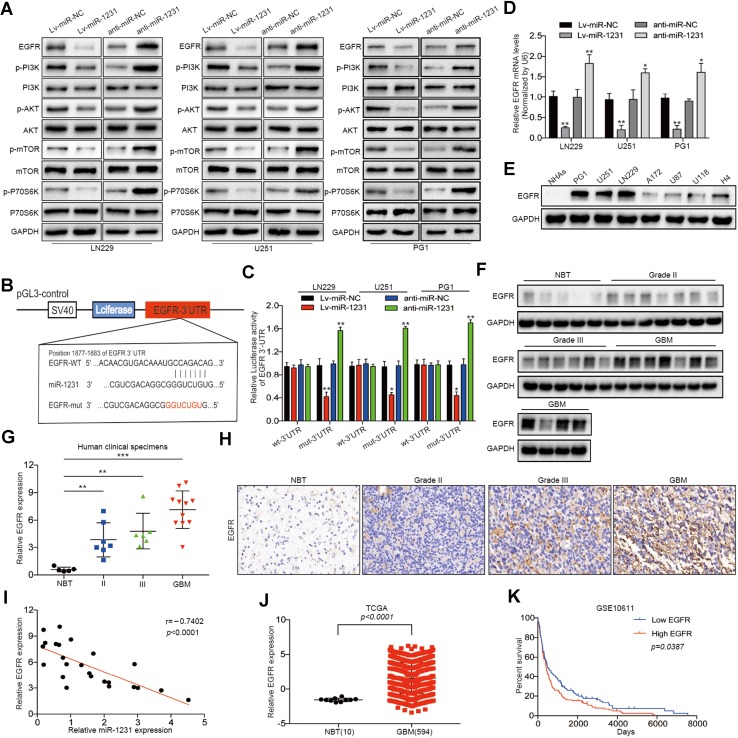
Fig. 3.
miR-1231 directly targets EGFR and inhibits the PI3K/AKT signaling pathway. a Western blot analysis of the indicated proteins in LN229, U251, and PG1 cells 48 h after transfection with miR-1231 lentivirus infection or anti-miR-1231. Data are presented as the means of triplicate experiments. b Schematic of the luciferase construct with the EGFR 3′ UTR containing a miR-1231 binding sequence. The seed sequence is shown in red font. Alignment was performed with the TargetScan 7.0 bioinformatics algorithm. c Luciferase reporter assays in cells co-transfected with the pGL3-EGFR wt-3′ UTR or pGL3-EGFR Mut-3′ UTR reporter and the indicated constructs (miR-NC, miR-1231, anti-miR-NC, or anti-miR-1231). Renilla luciferase vector was used as an internal control, **P < 0.01. d Relative EGFR mRNA expression levels in LN229, U251, and PG1 cells transfected as indicated were determined by qRT-PCR. U6 served as an internal control, **P < 0.01. e Western blot analysis of EGFR expression in glioma cell lines (PG1, LN229, U251, A172, U87, U118, and H4) and NHAs. GAPDH served as an internal loading control. f, g Immunoblotting analysis of EGFR expression in normal brain tissues (NBTs) (n = 5) and grade II (n = 7), grade III (n = 6), and GBM (n = 11) tissue specimens. Expression levels were normalized by GAPDH. Data represent the means ± SD from three independent experiments, **P < 0.01, ***P < 0.001. h Representative immunohistochemistry staining showing upregulation of EGFR expression in human glioma tissue specimens (clinical stage II–IV) compared with NBT. i Correlation between EGFR and miR-1231 expression levels in 24 human glioma tissues was analyzed by Spearman correlation (Spearman correlation analysis, r = − 0.7402, P < 0.001). j Analysis of the TCGA public database indicated that EGFR expression was significantly increased in GBM tissues compared with non-cancerous tissues, P < 0.001. k Kaplan–Meier curves showing the correlation in EGFR expression and overall survival of glioma patients using the GSE10611 database (P = 0.0387)