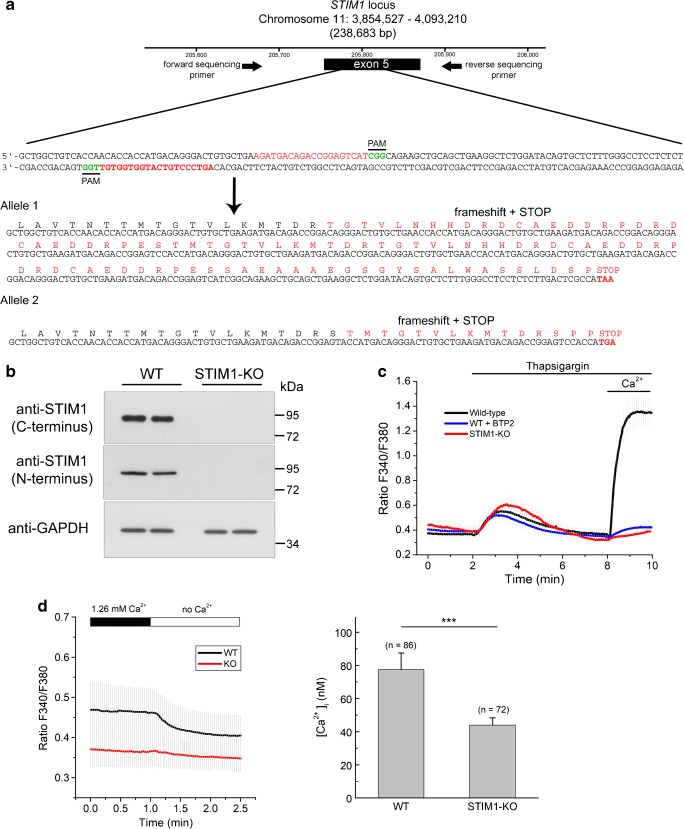
Fig. 3.
Knockout of STIM1 expression by CRISPR/Cas9 D10A gene editing. a Strategy for gene editing using CRISPR/Cas9 D10A in SH-SY5Y cells. A pair of guide RNAs (sense and antisense) was designed to trigger a double nick at exon 5 of the STIM1 locus. PAM sequences are denoted in green font. Sequencing of a PCR product from the genomic DNA of the selected clone revealed a 211 + 318 base-pair insertion at the target site. The translated protein sequence is denoted in red font, with premature stop codons at the end of the sequences of both alleles. b The selected clone of cells was assessed for STIM1 expression by immunoblot, using two different anti-STIM1 antibodies generated against C-terminal and N-terminal epitopes. Anti-GAPDH antibody was used as loading control. c Ca2+ entry was assessed as in Fig. 2, i.e., triggering the emptying of intracellular stores with 1 μM Tg in Ca2+-free HBSS and adding 2 mM Ca2+ back to the medium after store emptying. When required, the SOCE inhibitor BTP2 (3 μM) was added together with Tg. Data are presented as the mean ± s.d. of three independent experiments (n = 85 cells for KO; n = 75 cells for wild-type; n = 42 cells for WT + BTP2). d Steady-state cytosolic free Ca2+ concentration in WT and STIM1-KO cells. Left panel: Fura-2-loaded cells were incubated in Ca2+-containing HBSS (1.26 mM), and [Ca2+]i was measured as indicated in the Methods section. After recording the F340/F380 ratio signal, the medium was replaced by Ca2+-free HBSS to evaluate the contribution of extracellular Ca2+ entry to the [Ca2+]i in resting conditions. Right panel: After calibration of the fura-2 signal, the [Ca2+]i in resting conditions in Ca2+-containing HBSS was 77.4 ± 10 nM for wild-type cells, and 44 ± 4.4 nM in STIM1-KO cells. Data are the mean ± s.d. of three independent experiments (n = number of cells for each condition)