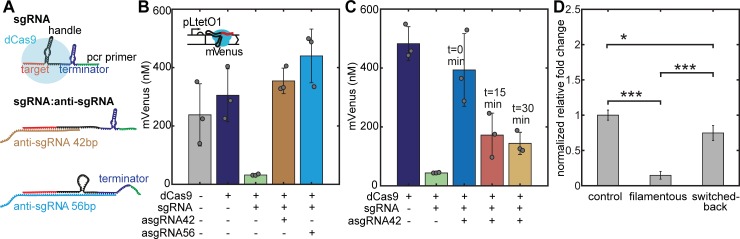
Fig 2. Experimental validation of the anti-sgRNA strategy.
(A) Secondary structures of free and complexed sgRNA and anti-sgRNA variants. The dCas9 handle is destroyed by duplex formation, which prevents dCas9 from binding. (B) Prototyping of the CRISPRi knockdown and rescue system in a cell-free gene expression system using mVenus as a fluorescent reporter protein. The restoration of mVenus expression is shown with truncated anti-sgRNA versions (the number indicates the number of base pairs in the resulting sgRNA:anti-sgRNA duplex). The expression of mVenus ([template DNA] = 5 nM) is blocked by the supplementation of purified dCas9 (70 nM) and sgRNA (100 nM) and is re-activated upon addition of anti-sgRNA (0.5 μM). Fluorescence levels for three different samples are taken at t = 15.5 hours or t = 11 h. Error bars are plotted as SD from 3 individual replicates. (C) Delaying the time of anti-sgRNA addition relative to dCas9-sgRNA results in lower mVenus fluorescence intensities. (taken at t = 12 hours). Error bars are plotted as SD from 3 individual replicates (c(sgRNA) = 250 nM, c(anti-sgRNA) = 1 μM). (D) Bar plot of RT-qPCR data, showing ftsZ RNA levels of normal, filamentous (107 nM, 1 mM IPTG) and switched back (107 nM, 1 mM IPTG, 100 nM AHL) cells from three technical replicates. The asterisk marks indicate the significance levels between the samples.