Figure 4.
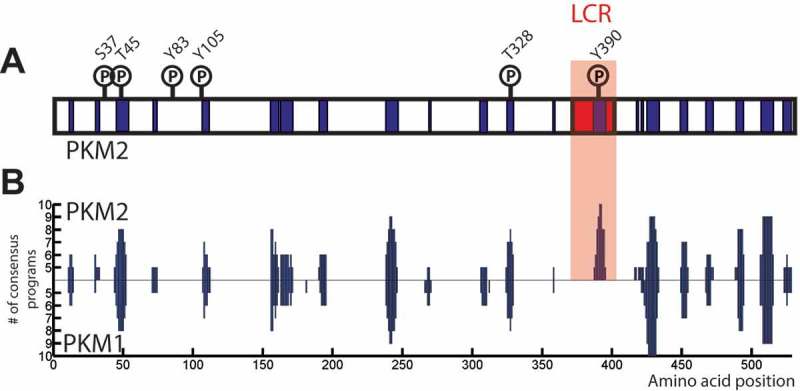
The mammalian pyruvate kinase PKM2 might also form amyloid-like aggregates, which could be regulated similar to Cdc19. A-B) Both PKM1 and PKM2 contain several stretches of amino acids that are predicted to form amyloids. However, only PKM2 presents a predicted LCR, which overlaps with the region with highest fibrillation propensity (highlighted in the red box). In addition, known PKM2 phosphorylation sites show a remarkable overlap with predicted amyloidogenic regions, suggesting that PKM2 might aggregate in a regulated way similar to Cdc19. The fibrillation profiles of PKM2 (upper part) and PKM1 (lower part) were computed using AmylPred2.0 (http://aias.biol.uoa.gr/AMYLPRED2/) [57]. Predicted amyloidogenic regions are shown in in blue. PKM2 phosphorylation sites were found in the Phospho.ELM database (http://phospho.elm.eu.org) [79]. PKM2’s predicted low complexity region was identified by the SEG program (http://mendel.imp.ac.at/METHODS/seg.server.html) [67], using a trigger window length [W] = 25, trigger complexity [K(1)] = 3.0 and extension complexity [K(2)] = 3.3., PKM2 LCR is highlighted in red.
