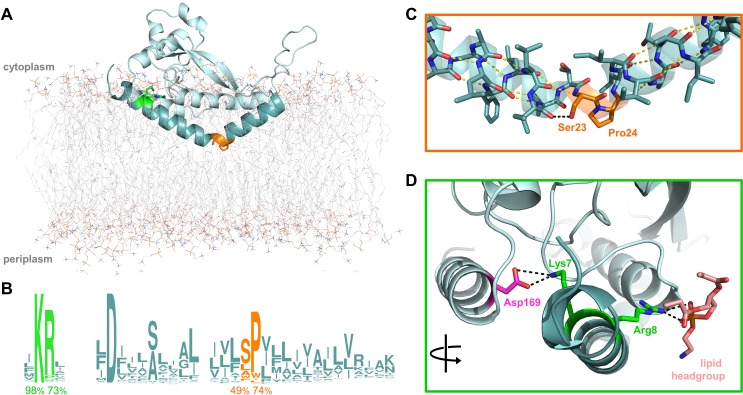
Figure 1. Overview of PglC highlighting the two conserved motifs in the RMH.
(A) Model of PglC from C. concisus in a POPE lipid bilayer. The RMH is shown in teal, Lys7 and Arg8 as green sticks, Ser23 and Pro24 as orange sticks. (B) Sequence logo showing conservation in the RMH domain among PglC homologs. Percent conservation is noted below each residue of interest. Logo generated using weblogo.berkeley.edu. (C) Detailed view of the Ser-Pro motif. Pro24 disrupts the hydrogen-bonding network (yellow dashes) of the RMH backbone. A 2.6 Åhydrogen bond (black dashes) is formed between the hydroxyl side group of Ser23 and the backbone carbonyl of Ile20. (D) Detailed view of the Lys-Arg motif. Lys7 forms a salt bridge with Asp169 (magenta). Arg8 interacts with a the headgroup of a copurified lipid (salmon).