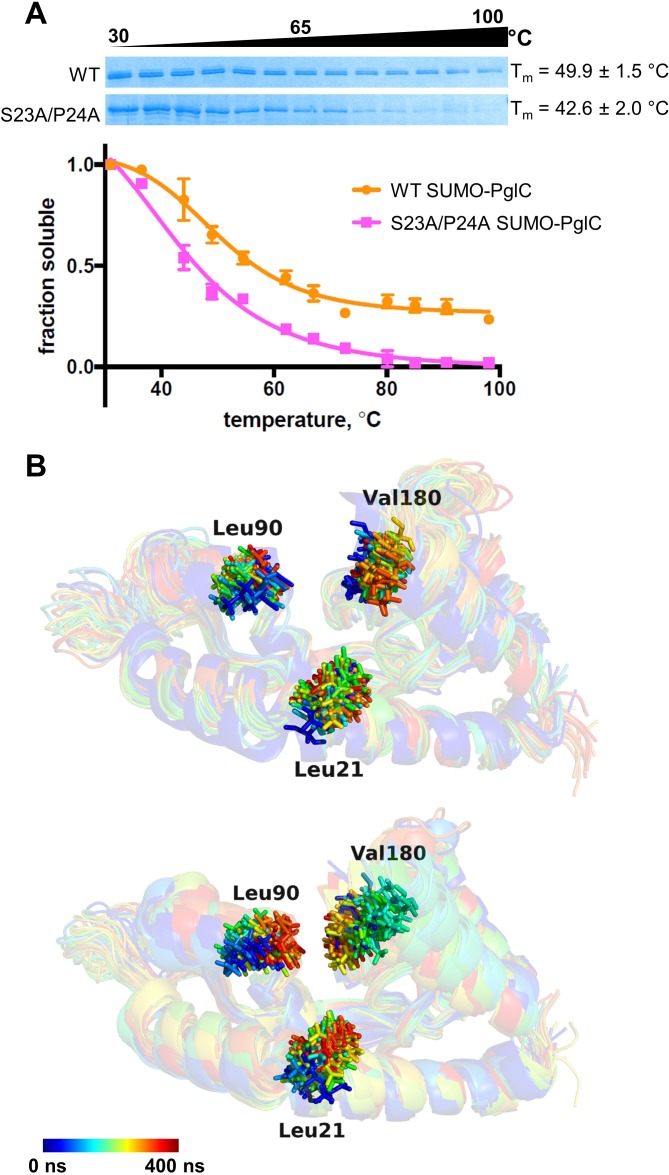
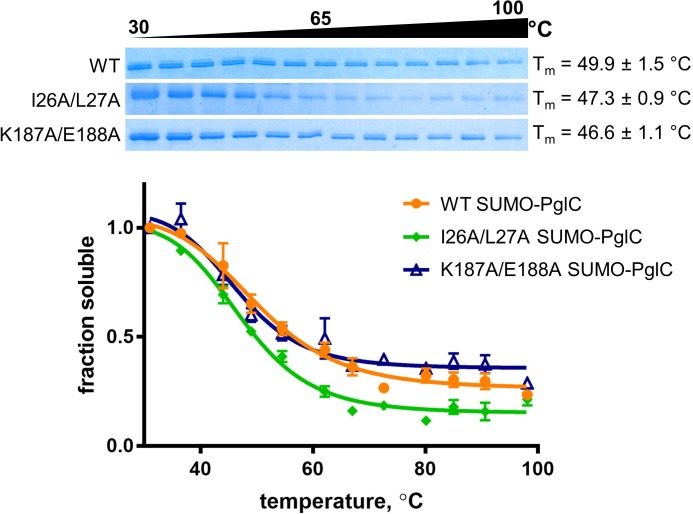
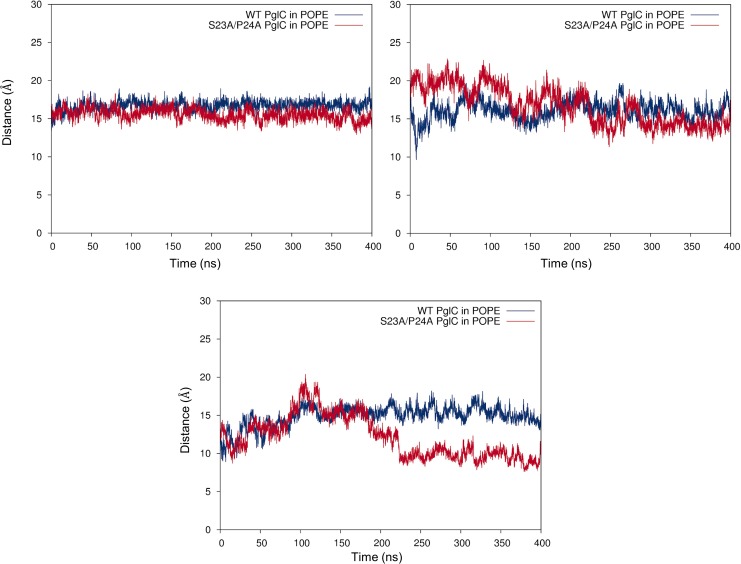
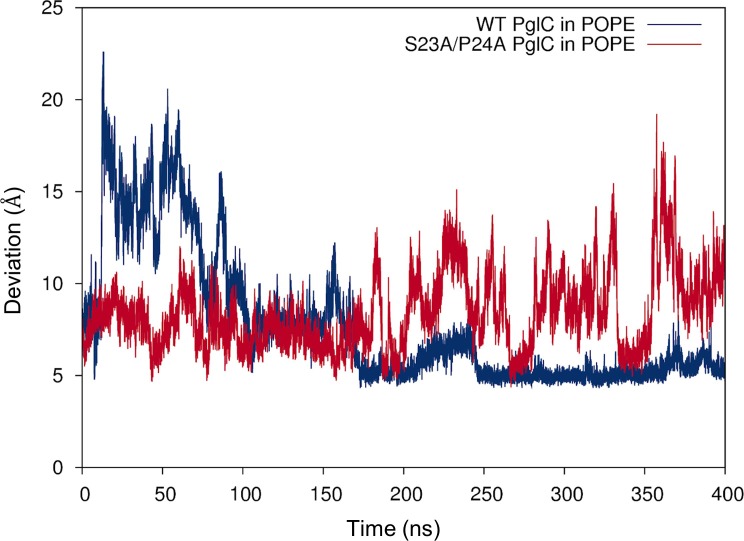
Figure 4. The Ser-Pro motif contributes to stability of the PglC fold.
(A) Thermal shift analysis of wild-type and S23A/P24A SUMO-PglC. Error bars are given for mean ± SEM, n = 3. (B) Superimposition of frames, taken at 10 ns intervals, along MD simulations of wild-type (top panel) and S23A/P24A (bottom panel) PglC. Colored from blue, t = 0 ns to red, t = 400 ns. PglC is represented as a semi-transparent cartoon and residues Leu21, Leu90 and Val180 as sticks.
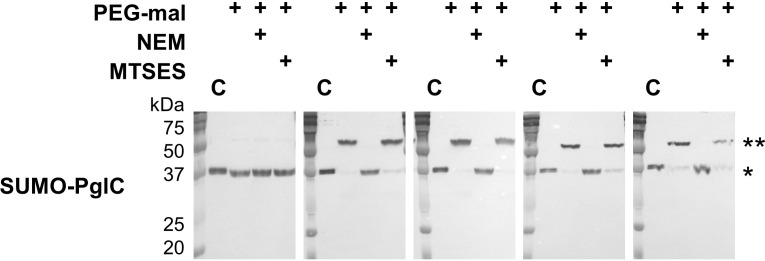
Figure 4—figure supplement 1. SUMO-PglC shows a reentrant topology similar to native PglC.
Figure 4—figure supplement 2. SDS-PAGE analysis of wild-type and S23A/P24A SUMO-PglC.