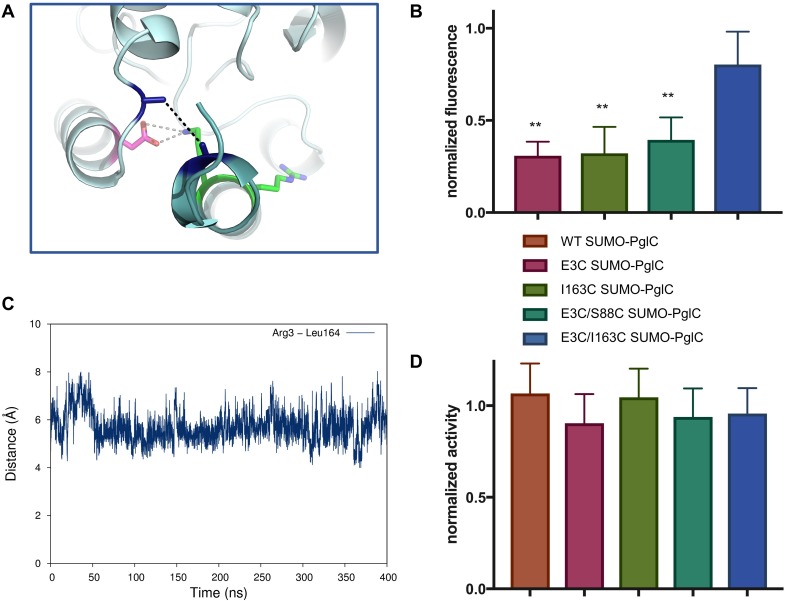
Figure 6. The RMH is held in the observed conformation during catalysis.
(A) Detailed view showing the location of bBBr crosslinking (black dashes). The Cα and Cβ of Arg3 and Leu164 in the structure of PglC from C. concisus are shown as dark blue sticks (the remainder of the side chains is omitted for clarity). The corresponding residues Glu3 and Ile163 in PglC from C. jejuni were substituted with Cys for bBBr crosslinking studies. (B) Fluorescence of SUMO-PglC variants following crosslinking with bBBr, normalized to fluorescence of DTT-quenched samples (quenched samples represent maximum possible fluorescence). Error bars are given for mean ± SD, n = 4 (**p<0.01, Student’s t-test; p-values for each variant are 0.0022 (E3C), 0.0055 (I163C), 0.0091 (E3C/S88C). (C) Distance between Arg3 and Leu164 (measured from the centroid of each residue), over 400 ns of MD simulations of PglC in a POPE membrane. (D) Activity of SUMO-PglC variants following crosslinking with bBBr, normalized to activity following treatment with vehicle. Error bars are given for mean ± SD, n = 3.