Fig. 1.
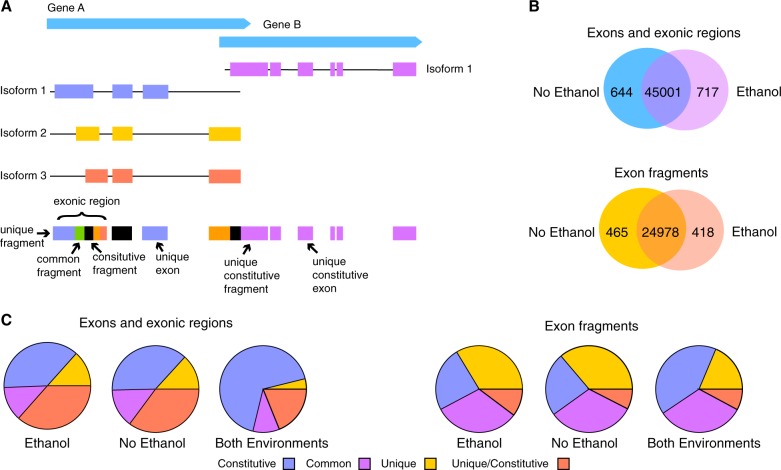
a A representation of the classification scheme for exons, exonic regions, and exon fragments. In higher eukaryotes exons from different isoforms of a gene may overlap, and portions may be shared between some or all isoforms. In effect, this makes it essentially impossible to disentangle expression and splicing, and hence we classify exons as either being entire exons, which do not overlap other exons in different isoforms (exons), or fused regions consisting of a set of overlapping exons (exonic regions). Exonic regions can be decomposed into exon fragments, depending upon their overlap between different isoforms. Exons may be unique (found in a single isoform), common (found in some isoforms but not all), or constitutive (present in all isoforms), and if they are unique to a single isoform they may also be unique and constitutive if that is the only transcript annotated for that particular gene. As exonic regions require overlap between exons they may not be unique, but they may be common or constitutive. Exon fragments can be unique to a single isoform, common to several isoforms, or constitutive. Exon fragments may be unique and constitutive only in the situation of a multi-gene exonic region where one gene has a single isoform. b Some exons, exonic regions, and exon fragments could not be compared for expression levels for treatment because they were only unambiguously detected in one environment. Shown here is the number of exons, exonic regions, and exon fragments detected without ethanol, with ethanol, and in both environments. c The proportion of exons, exonic regions, and exon fragments that were unique, unique/constitutive, common, or constitutive and were detected either only without ethanol, only with ethanol, or in both environments. As expected constitutive exons and exonic regions are more commonly detected in both environments, which is suggestive of alternative exon usage in response to ethanol