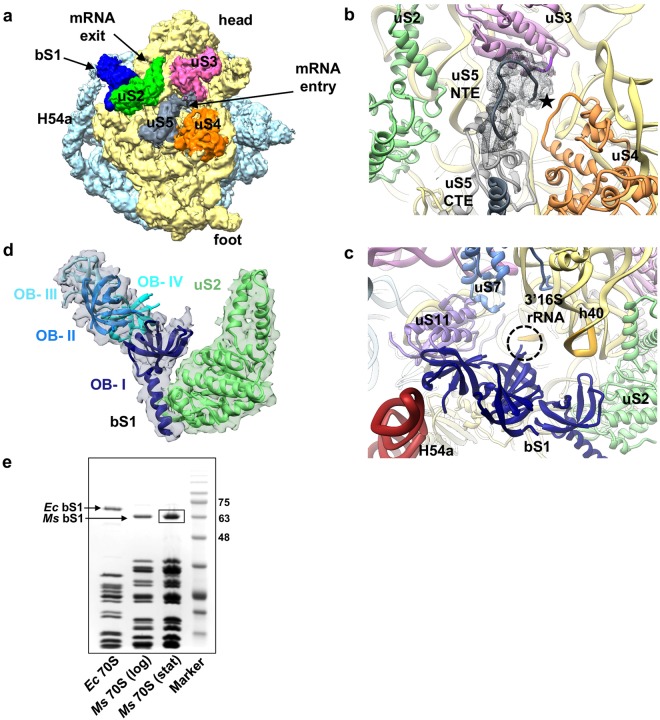
Figure 2.
Architecture of mRNA entry and exit sites in the hibernating state of Ms 70S ribosome. (a) A surface view of the SSU in hibernating state of 70S shown from the solvent-side. Structural landmarks of SSU and the proteins around the mRNA entry and exit sites are labelled as indicated. The density for bS1 is shown at threshold 0.025 and for the rest at threshold 0.04 in UCSF chimera55. (b) mRNA entry site in Ms is formed by three r-proteins, uS3, uS4, and uS5. The uS5 NTE extends towards the mRNA entry channel (marked with a star symbol) and CTE forms an α-helix on the SSU surface. Density of uS5 NTE is shown in grey mesh. (c) mRNA exit site (marked by a dotted circle) in Ms is formed by two r-proteins uS7 and uS11, and 16S rRNA. bS1 protein is present near to the mRNA exit. The N-terminal α-helix of Ms bS1 interacts with uS2 and one of the OB domain (domain III) with H54a. bS1 is colored in deep blue, uS2 in green, uS3 in pink, uS4 in orange, and uS5 in grey colors in (a–c). (d) Zoomed-in view of the bS1. Density for bS1 is in transparent grey, the four OB domains of bS1 are labelled as indicated and highlighted in different shades of blue. Densities for OB domains and uS2 protein are shown at the different thresholds for clarity. (e) 15 pmol of 70S ribosomes from Ec, Ms (log phase), and Ms (stationary phase) were analysed on 12% SDS-PAGE with coomassie blue staining. The bS1 protein is highlighted. Ms 70S (stationary phase) bS1 protein band (boxed) is further analysed by mass spectrometry (Supplementary Fig. S9).