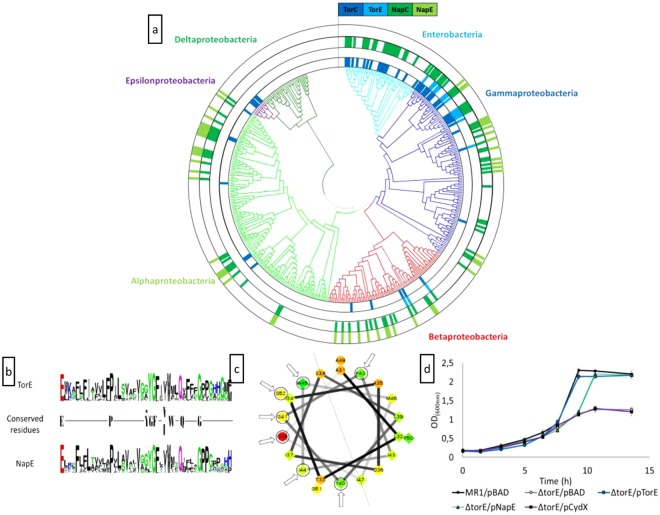
Figure 1.
(a) Occurrence of torC, torE, napC and napE genes among the proteobacteria. The presence of each gene is represented by block coloured in dark blue, light blue, dark green and light green, respectively. The phylogenetic tree has been constructed using the GyrB protein sequence from a bacterium of each genus of proteobacteria. Branches are coloured according to the class of proteobacteria. Within the gammaproteobacteria class, the enterobacteria order was specifically indicated. (b) Sequence logo of the TorE and NapE proteins. The conserved residues in the two sequences are shown. The black star indicated that the tyrosine residue is highly conserved in the NapE proteins except in bacteria that also possess the TorE protein. (c) Helical projection of the hydrophobic stretch of the TorE protein of S. oneidensis. These residues (from 31 to 52) were selected because of their location between two prolines. The conserved residues are indicated by arrows. The centreline of the helix is represented. (d) Complementation experiments. Growth curves of the wild type strain (MR1/pBAD) and ΔtorE mutant harbouring either pBAD, pTorE, pNapE or pCydX plasmid. Cells were grown anaerobically in LB medium in the presence of TMAO, arabinose and chloramphenicol. Each point represents the average ± standard error of three independent experiments.