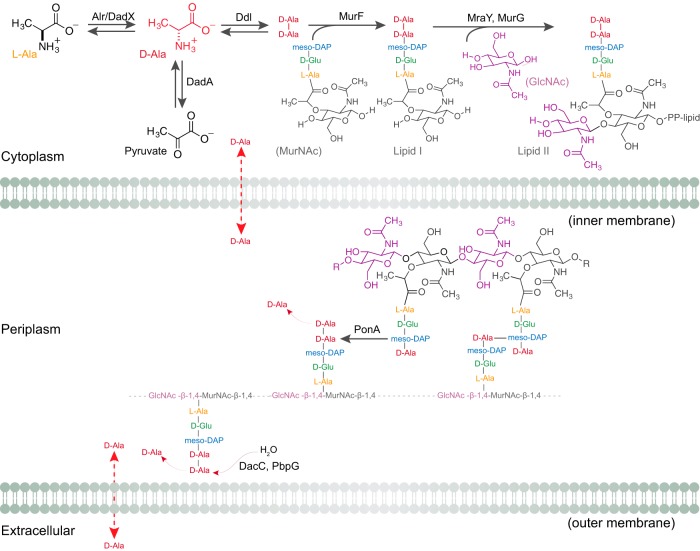
FIG 1 .
Biochemistry of d-Ala in Gram-negative bacteria. The cartoon represents the utilization and role of d-Ala in bacterial cells. P. aeruginosa cells have two alanine racemases (Alr and DadX) that interconvert l-Ala and d-Ala. DadA is a d-amino-acid dehydrogenase that degrades d-Ala into pyruvate. Ddl is an amino acid ligase that converts two d-Ala molecules into d-Ala-d-Ala, which is a substrate of the enzyme MurF in forming lipid I from the MurNAc tripeptide. MraY and MurG form lipid II, which is subsequently flipped across the membrane into the periplasm and incorporated into the growing peptidoglycan. The PonA transpeptidase cross-links stem peptides during peptidoglycan biosynthesis by releasing the terminal d-Ala into the periplasm. dd-Carboxypeptidase (DacC) and dd-endopeptidases (PbpG) also release the terminal d-Ala from the un-cross-linked lipid II in the periplasm. Free d-Ala in the periplasm and in the extracellular environment is transported into cells through alanine transporters and permeases. “PP-lipid” refers to a diphosphate bridge and long, connected hydrocarbon tail that is attached to the disaccharide in lipid II.