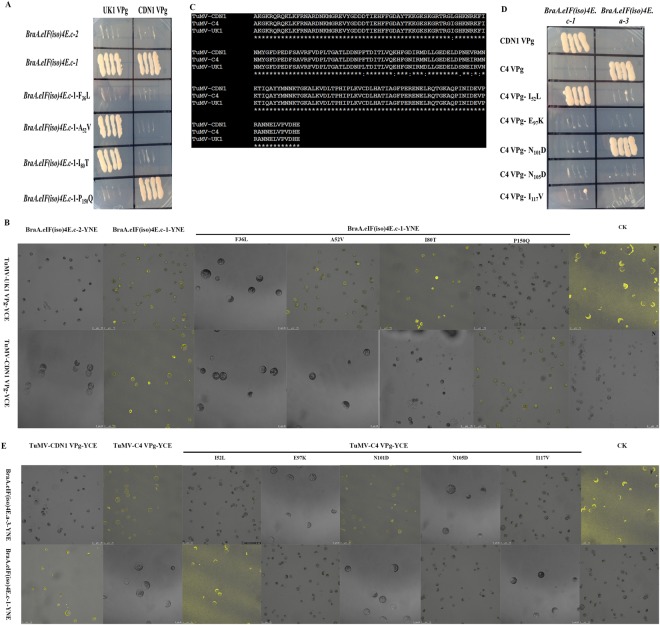
Figure 3.
Specific SNPs affect the interaction between eIF(iso)4E and TuMV VPg. Variations between BraA.eIF(iso)4E.c-1 and BraA.eIF(iso)4E.c-2 could affect the interaction as confirmed by Y2H (A) and by BiFC. (B,C) Multiple sequence alignment of TuMV C4 VPg TuMV UK1 VPg and TuMV CDN1 VPg. Five amino acid substitutions were identified between TuMV C4 and CDN1, whereas four amino acid substitutions were detected between TuMV C4 and UK1. Variations between TuMV-C4 VPg and TuMV-CDN1 VPg could affect the interaction as indicated by Y2H (D) and by BiFC. (E) Y2H: negative control, the empty vectors pGADT7 and pGBKT7 (data not shown); positive controls, the murine p53 and SV40 large T antigen from the Matchmaker GAL4 two-hybrid system 3; TuMV-VPg and Arabidopsis eIF(iso)4E (lsp); assay controls: each partner and empty vector (data not shown). BiFC: P-positive controls (the combination of bZIP63YN and bZIP63YC); N-negative controls (YNE-empty and YCE-empty vectors); the assay controls were each partner and empty vectors (data not shown).