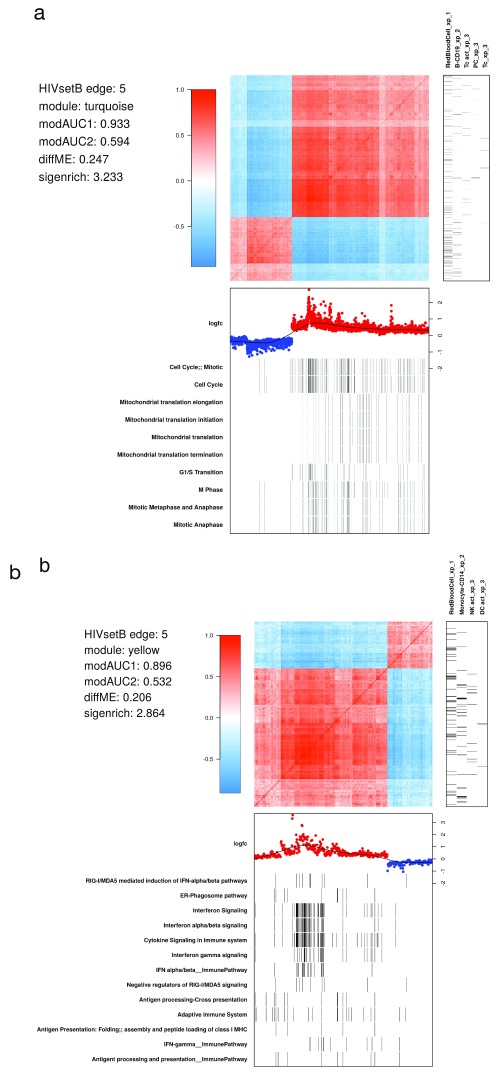
Figure 6. WGCNA module structure.
( A) Correlation matrix of all probes in the turquoise module in the HIVsetB dataset (N HIV=30, N Controls=17, see Figure S1; Supplementary File 1). Colours in the heatmap represent Pearson correlation coefficients, ranging from -1 to 1, as indicated by the legend. The module is enriched for lymphocyte-specific genes (right annotation panel) as well as cell cycle/mitosis associated genes, suggesting that various lymphocyte subsets in acute HIV infection are actively proliferating. (bottom annotation panel). Log 2-fold change values refer to differential transcript abundance in acute HIV relative to healthy controls. ( B) Correlation matrix of all probes in the yellow module in the HIVsetB dataset. It is enriched for innate cell genes as well as interferon signaling, suggesting that innate immune cells are in an interferon-induced state. Additional annotation information is provided to the left of the heatmap. The parameters modAUC1, modAUC2, diffME and sigenrich are defined in Supplementary methods. The plot is generated using a custom R function ( mwat).