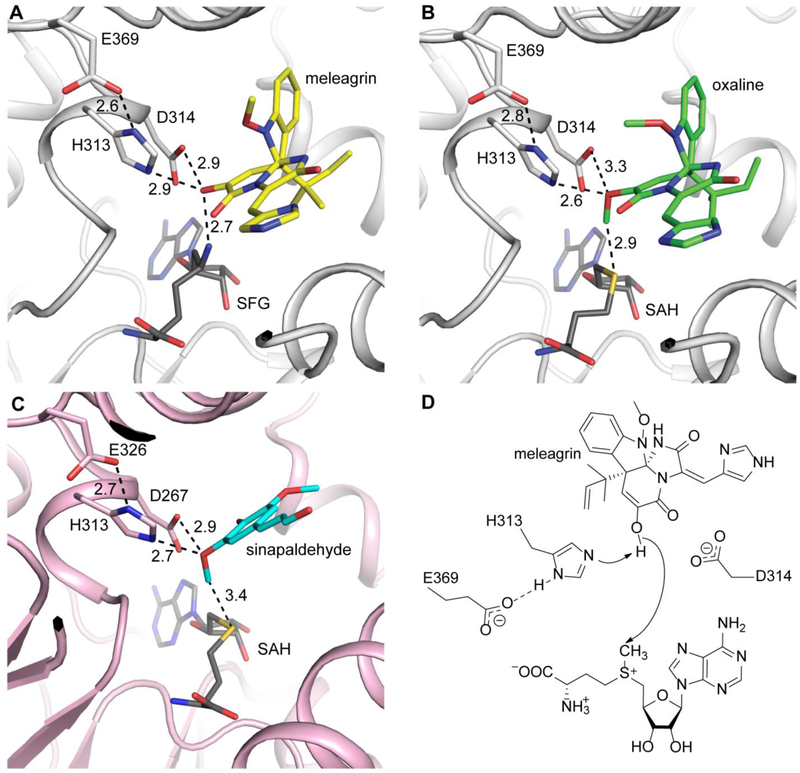
Fig. 7. OxaC active site architecture and mechanism.
(A) Meleagrin (yellow) is ideally positioned for SN2 attack of the donor methyl. (B) The product oxaline (green) shows little displacement after methylation. (C) CoMT (pink) shows homologous active site organization. The phenolic acceptor sinapaldehyde (cyan) is similarly activated by His and Asp residues. All distances are labeled in Ångstroms. (D) Proposed OxaC mechanism. Glu369 primes the active site base His313 for deprotonation of the enol. Asp314 likely plays a role in acceptor positioning.