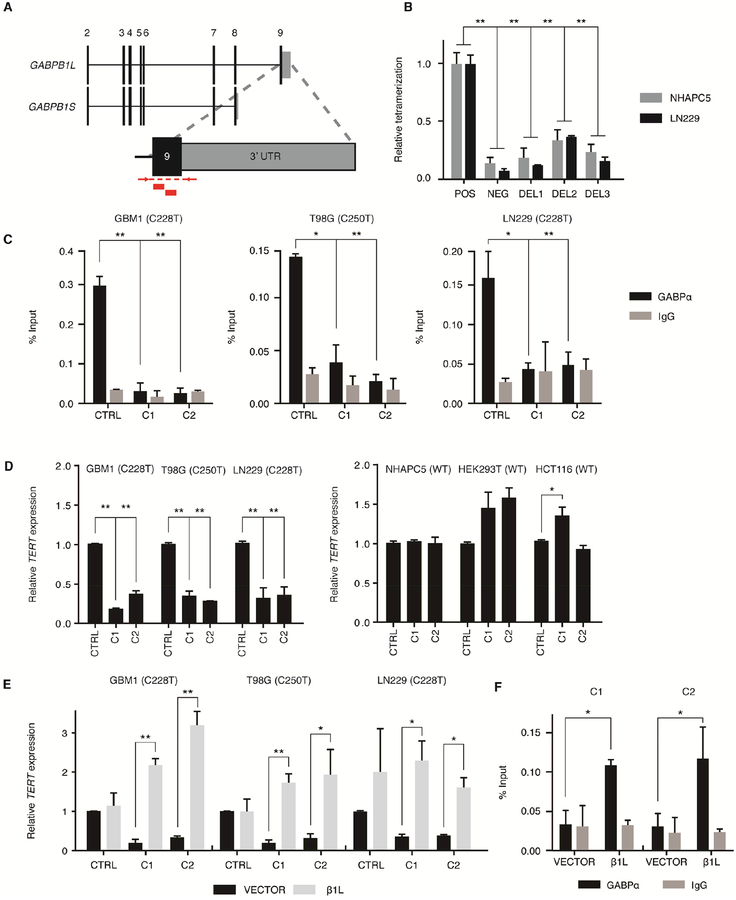
Figure 2.
CRISPR-Cas9-mediated disruption of GABPB1L reduces GABP-mediated activation of the mutant TERT promoter. (A) Exon structure for the GABPB1 locus, depicting the GABPB1S and GABPB1L isoforms. Inset shows targeting strategy for CRISPR-Cas9 editing of GABPB1L. Red blocks indicate sgRNA target sites. Red arrows and dashed lines indicate primer locations and target amplicon for PCR validation of editing. (B) Quantification of β1L tetramerization in the wild-type (POS) or mutated (DEL1-3) state. The negative (NEG) state consists of one β1L vector and one β1S vector, the products of which are unable to form a tetramer. *p value<0.05, **p value<0.01, two-sided Student’s t-test of DEL1-3 or NEG respective to the positive control (POS). (C) GABPα or IgG control ChIP-qPCR for the TERT promoter in CRISPR control (CTRL) or β1L-reduced clones (C1 and C2). *p value<0.05, **p value<0.01, two-sided Student’s t-test compared to respective CTRL. (D) TERT expression relative to CTRL for β1L-reduced TERT promoter mutant (left) or wild-type (right) clones. *p value<0.05, **p value<0.01, two-sided Student’s t-test compared to CTRL. (E,F) TERT expression (E) or GABPα occupancy (F) in β1L-reduced clones relative to CTRL 48 hr following transfection with empty (VECTOR) or β1L expression vector. *p value<0.05, **p value<0.01, two-sided Student’s t-test compared to respective VECTOR control. Values are mean ± S.D. of at least two independent experiments (C and F) or three independent experiments (B, D, and E). See also Figures S2–S3 and Tables S1–S3.