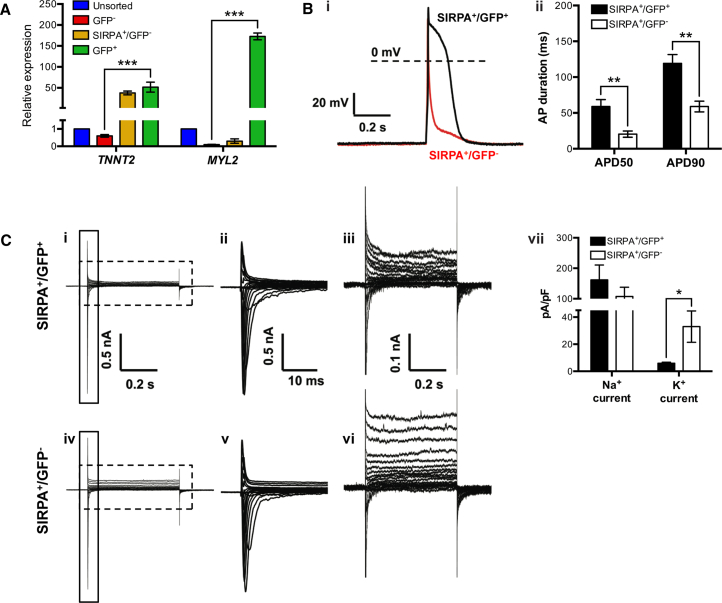
Figure 2.
Characterization of H9 MYL2-GFP hESC-Derived Ventricular Cardiomyocytes
(A) qRT-PCR analysis of TNNT2 and MYL2 gene expression in unsorted, MYL2-GFP-negative (GFP−)-, SIRPA-bright/MYL2-GFP-negative (SIRPA+/GFP−)-, and MYL2-GFP-positive (GFP+)-sorted fractions of dissociated hESC-derived embryoid bodies at day 25 of cardiac differentiation (see Figure S2 for cell gating strategy). Data were normalized to corresponding ACTB expression, and are relative to the unsorted hESC-derived cell population. Error bars, mean ± SD. ∗∗∗p ≤ 0.001 by Student's t test (n = 3 technical replicates). Data are representative of a minimum of three independent biological replicates.
(B) Electrophysiological characterization of SIRPA+/GFP+- and SIRPA+/GFP−-sorted fractions of dissociated hESC-derived embryoid bodies at days 25–40 of cardiac differentiation (see Figure S2B for cell gating strategy). (i) Representative triggered action potential (AP) traces recorded from SIRPA+/GFP+ and SIRPA+/GFP− cardiomyocytes using whole-cell current-clamp technique. (ii) Quantification of AP duration (APD) at different times of repolarization (50% and 90%) for SIRPA+/GFP+ (n = 21) and SIRPA+/GFP− (n = 18) cardiomyocytes. Error bars, mean ± SEM. ∗∗p ≤ 0.01 by Student's t test. mV, millivolts; s, seconds. Data are representative of a minimum of three independent biological replicates.
(C) Representative whole-cell currents recorded from (i) SIRPA+/GFP+ (upper panel) and (iv) SIRPA+/GFP− (lower panel) cells, with an enlarged view of the solid boxed area (ii and v), and the dashed boxed area (iii and vi) to the right of the corresponding trace. (vii) Quantification of maximum inward Na+ currents, and maximum sustained K+ currents of SIRPA+/GFP+ (n = 21) and SIRPA+/GFP− (n = 18) cardiomyocytes. Error bars, mean ± SEM. ∗p ≤ 0.05 by Student's t test. Data are representative of a minimum of three independent biological replicates. nA, nanoampere; s, seconds; ms, milliseconds; pA/pF, picoampere per picofarad.