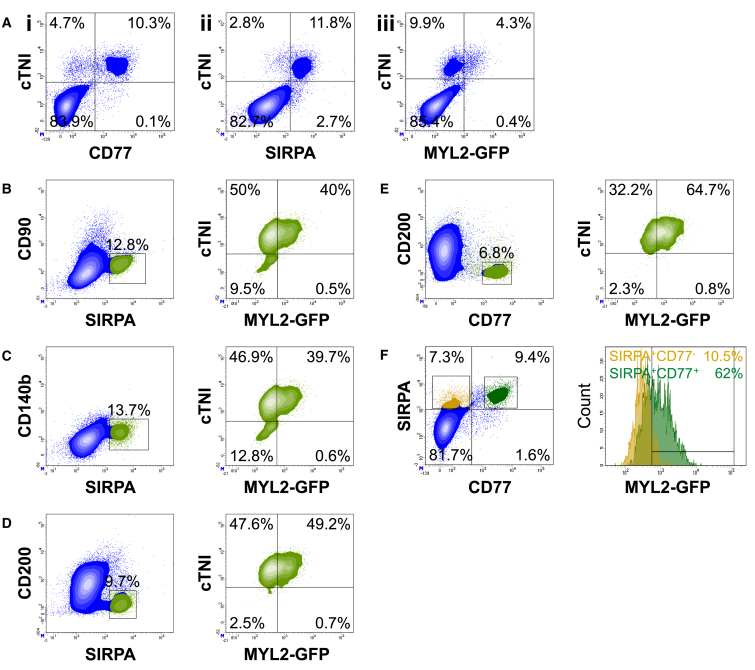
Figure 5.
CD77 Expression Highly Correlates with hESC-Derived Ventricular Cardiomyocyte Phenotype
All analysis is of the same population. Data are representative of a minimum of three independent biological replicates.
(A) Correlation of cTNI and (i) CD77, (ii) SIRPA, or (iii) MYL2-GFP in hESC-derived populations at day 25 of cardiac differentiation by flow cytometry.
(B–E) Representative plots depicting cTNI/MYL2-GFP cell populations (right panel, green plots) at day 25 of cardiac differentiation, after gating on different cell populations defined by distinct surface marker signatures: (B) SIRPA+/CD90−; (C) SIRPA+/CD140b−; (D) SIRPA+/CD200−; and (E) CD77+/CD200− (left panel, boxed area).
(F) Left panel: SIRPA+/CD77− (yellow) and SIRPA+/CD77+ (green) cell populations were defined within single cells. Right panel: histograms depicting MYL2-GFP expression within SIRPA+/CD77− and SIRPA+/CD77+ cell populations.