Figure 6.
Validation of Ventricular Cardiomyocyte Enrichment from hESC-Derived Populations with a Cell-Surface Signature
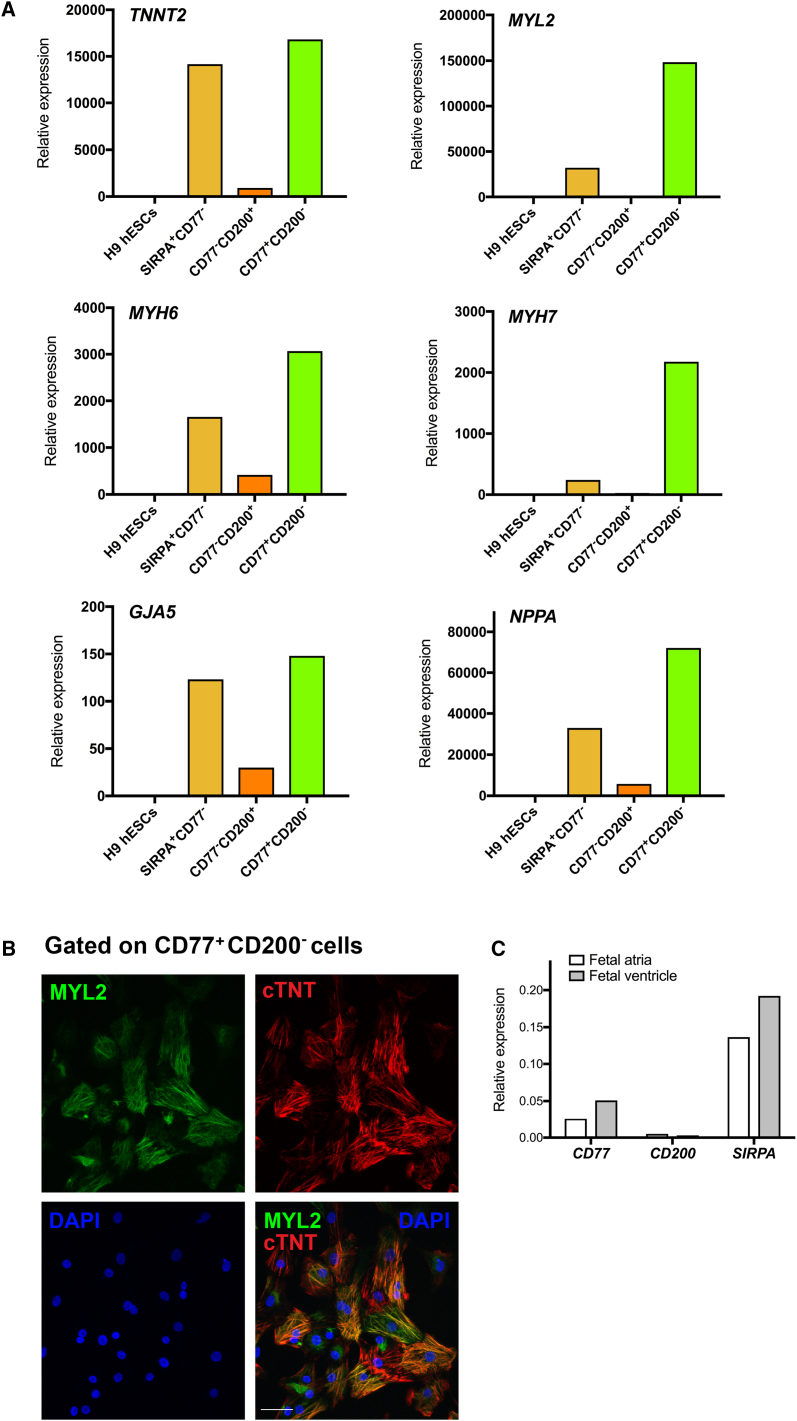
(A) qRT-PCR analysis of TNNT2, MYL2, MYH6, MYH7, GJA5, and NPPA gene expression in SIRPA+/CD77−-, CD77−/CD200+-, and CD77+/CD200−-sorted fractions of dissociated hESC-derived embryoid bodies at day 25 of cardiac differentiation. Data were normalized to corresponding ACTB expression, and are relative to H9 hESC-derived embryoid bodies at day 0. Data are representative of a minimum of three independent biological replicates.
(B) Immunofluorescence analysis of H9 hESC-derived CD77+/CD200−-sorted cells after 25 days of cardiac differentiation. Cardiomyocytes were stained with cTNT. DNA was stained with DAPI. Scale bar, 50 μm. Images are representative of a minimum of three independent biological replicates.
(C) qRT-PCR analysis of CD77, CD200, and SIRPA gene expression in human 15-week fetal atrial and ventricular tissue. Data were normalized to corresponding GAPDH expression.