FIGURE 2. CellTrace labeling of primary OT-I T cells allows for identification of OT-I nuclei after coculture.
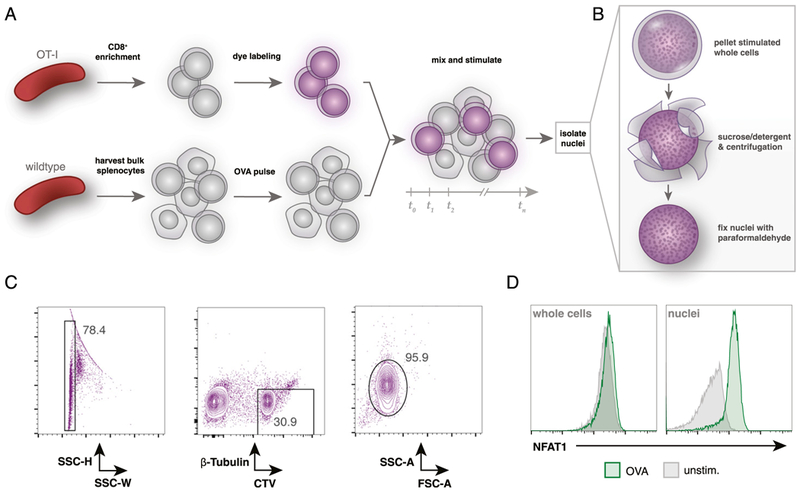
(A) Overview of coculture experimental setup. OT-I splenocytes are enriched for CD8+ T cells by negative selection and then labeled with CellTrace Violet dye. Wild-type splenocytes are harvested separately and pulsed with OVA peptide. Labeled OT-I cells are mixed (~1:3) with bulk splenocytes for desired time. (B) Summary of the nuclei isolation protocol. After coculture stimulation, all mixed OT-I and wild-type cells are lysed in a sucrose and detergent buffer, centrifuged, and washed twice prior to fixation with 4% paraformaldehyde. (C) Flow cytometry gating strategy to identify OT-I nuclei after coculture stimulation. Fixed nuclei are stained with a fluorescent Ab against β-tubulin prior to analysis. OT-I nuclei are identified by gating on singlets by side scatter height (SSC-H) and SSC width (SSC-W) and then on β-tubulinlo and CellTrace Violethi events. (D) Flow cytometry comparison of isolated OT-I nuclei or whole cells (prelabeled with CellTrace Violet) after 30 min of coculture with 1 nM OVA peptide-pulsed bulk C57BL/6 splenocytes. Gated on CellTrace Violethi and β-tubulinlo events.
