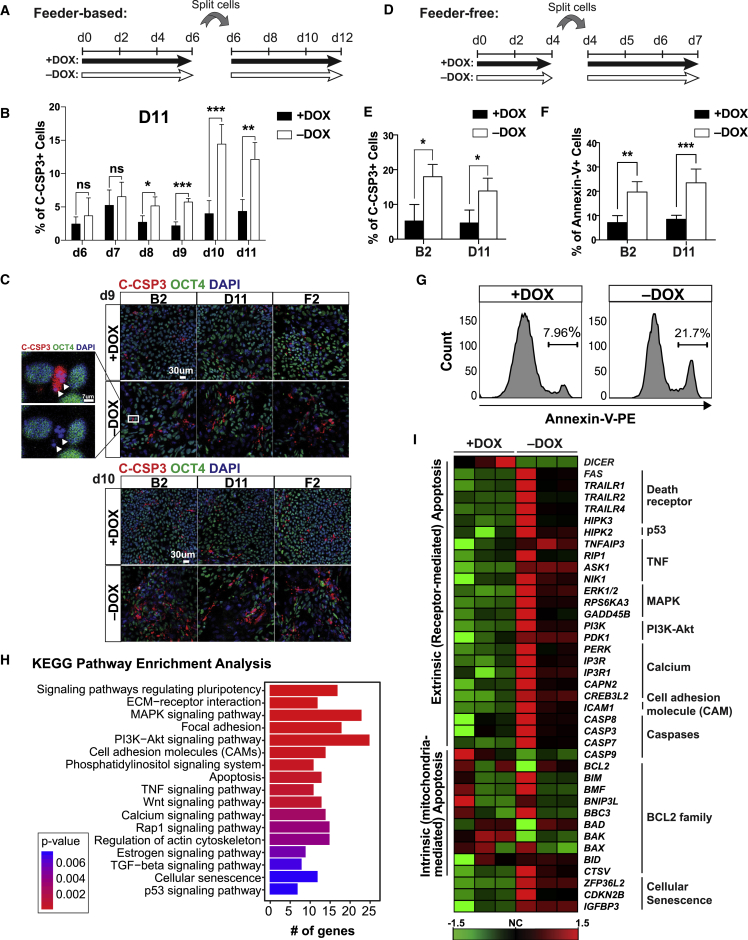
Figure 3.
DICER1 Depletion in hESCs Causes Caspase-3-Mediated Apoptosis
(A) Schematic representation of the experimental design in feeder-based condition. Cells were split at 28,000 cells/cm2. DOX, doxycycline.
(B) Flow cytometry of cleaved caspase-3 (C-CSP3) from days 6 through 11 in the D11 conditional DICER1 knockout line with and without DOX in feeder-based condition. n = 3 independent experiments.
(C) Representative immunofluorescence pictures of C-CSP3 and OCT4 in the D11 conditional DICER1 knockout line maintained with and without DOX at days 9 and 10 in feeder-based condition. Inset represents a close-up view showing fragmentation (arrowheads) of nuclei and loss of OCT4 in a C-CSP3-positive cell. Scale bars, 30 μm (main images) and 7 μm (close-up). n = 3 independent experiments.
(D) Schematic representation of the experimental design in feeder-free condition. Cells were split at a density of 14,000 cells/cm2.
(E) Flow-cytometry detection of C-CSP3 expression in B2 and D11 conditional DICER1 knockout lines in feeder-free condition maintained with and without DOX for 7 days. n = 4 independent experiments.
(F) Flow cytometry of annexin-V in B2 and D11 conditional DICER1 knockouts cultured in feeder-free condition maintained with or without DOX for 7 days. n = 4 independent experiments for B2, and n = 5 independent experiments for D11.
(G) Representative histograms of annexin-V flow cytometry on D11 conditional DICER1 knockout line maintained with or without DOX in feeder-free condition. n = 5 independent experiments.
(H) KEGG pathway enrichment analysis was performed in R (v3.4.2) with clusterProfiler (v3.6.0) on RNA-seq data of B2 conditional DICER1 knockout line cultured in feeder-based condition with or without DOX for 9 days. n = 3 independent experiments.
(I) Heatmap of apoptosis-related genes. Data are derived from RNA-seq of B2 conditional DICER1 knockout hESCs with and without DOX for 9 days in feeder-based condition. The heatmap was generated by cluster analysis using the online analysis tool CARMAweb (https://carmaweb.genome.tugraz.at/genesis) with the methods normalized gene. The variation between the biological repeats could be due to the rapid nature of apoptosis. n = 3 independent experiments.
Error bars indicate SD, and significance is indicated as ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001; ns, not significant (p ≥ 0.05). See also Figure S3.